6.3. TSUNAMI Utility Modules
B. T. Rearden, M. A. Jessee, and J. D. McDonnell
ABSTRACT
Several modules have been developed to assist with the sensitivity and uncertainty analysis techniques included in SCALE. TSUNAMI-IP (Tools for Sensitivity and Uncertainty Analysis Methodology Implementation-Indices and Parameters) uses sensitivity data generated by TSUNAMI-1D, -2D, or -3D to propagate the effect of uncertainty in nuclear data to a response of interest, and to generate several relational parameters and indices that predict the degree of similarity between two systems. Additionally, the formats of files used by SCALE for sensitivity analyses are described herein.
ACKNOWLEDGMENTS
The author wishes to acknowledge C. M. Hopper, B. L. Broadhead of the Oak Ridge National Laboratory (ORNL), S. Goluoglu, and R. L. Childs, formerly of the ORNL, and J. J. Wagschal of The Hebrew University of Jerusalem, for their assistance with the development of TSUNAMI-IP and L. M. Petrie, I. C. Gauld, and J. E. Horwedel of the Oak Ridge National Laboratory for their assistance in the development of SENLIB and BONAMIST.
6.3.1. TSUNAMI-IP
TSUNAMI-IP (Tools for Sensitivity and Uncertainty Analysis Methodology Implementation-Indices and Parameters) uses sensitivity data generated by TSUNAMI-1D, -2D, or -3D and cross section covariance data described in the SCALE Nuclear Data Covariance Library chapter to generate several relational parameters and indices that can be used to determine the degree of similarity between two systems. TSUNAMI-IP combines many techniques developed over several years into a comprehensive package with simplified input. Most of the techniques developed for TSUNAMI-IP are documented in Ref. [refTsip1], other techniques are documented in Refs. [refTsip2], [refTsip3] and [refTsip4], and others are originally presented in this document.
Depending on the user selected options, voluminous output can be generated by TSUNAMI-IP. To assist with review of these data, TSUNAMI-IP was the first SCALE module to offer HTML formatted output. This document first describes the indices and parameters that are computed, presents a description of the user input, then finally a sample problem is presented where the standard text output and the HTML formatted output are described.
TSUNAMI-IP was introduced in SCALE 5.0 and improved in SCALE 5.1. In SCALE 6.0, TSUNAMI-IP now includes an individual ck to better quantify uncertainty-based similarity on a nuclide-reaction specific level. The Esum parameter has since been replaced with a sensitivity-only parameter, simply called E, which is based on more rigorous mathematical definition. The input format has been expanded with additional keywords to identify sensitivity data and a RESPONSE data block that allows users to define applications and experiments within the same input block. Uncertainty and penalty edits can also be presented with relative or absolute values, where previously only relative values were available. The input of user-defined covariance data now allows much greater flexibility, and the covariance data used in the problem can now be exported to a COVERX formatted data file for subsequent plotting in Fulcrum.
6.3.1.1. Global integral indices
Three global integral indices that assess the similarity of a particular application and a single experiment on a system-wide basis for all nuclides and reactions are defined in this section. The integral indices are ck, E, and G. Each of these indices is defined in subsequent subsections, and TSUNAMI-IP input options used to generate these indices and produce specific output edits are explained. Each integral index is normalized such that a value of 1.0 indicates complete similarity between the application and the experiment and a value of 0.0 indicates no similarity.
6.3.1.1.1. Integral index ck
A rigorous approach to assessing the similarity of two systems for purposes of criticality code validation is the use of uncertainty analysis, which propagates the tabulated cross section uncertainty information to the calculated keff value of a given system via the energy-dependent sensitivity coefficients.X1X Mathematically, the system uncertainty is computed with a quadratic product of the group-wise sensitivity profile vectors by nuclide and reaction type with the group-wise cross section uncertainty matrices by nuclide and reaction type. The result of this procedure is not only an estimate of the uncertainty in the system keff due to cross sections, but also an estimate of the correlated uncertainty between systems. These correlated uncertainties can be represented by correlation coefficients, which represent the degree of correlation in the uncertainties between the two systems. This correlation coefficient index, denoted as \(c_k\), not only has the desirability of a single quantity relating the two systems, but also measures the similarity of the systems in terms of related uncertainty. These correlation coefficients are particularly useful when used in traditional trending analyses for criticality safety validation in that the correlation coefficient relates the degree in which the uncertainties in the critical benchmarks are coupled with the uncertainties in the application of interest. This coupling with the common uncertainties in the various systems is expected to closely mimic the coupling in predicted biases between the various systems, since they should both be related to the cross section uncertainties.
The cross section covariance data are read from a COVERX formatted data file identified by coverx= in the PARAMETER input. The cross section covariance data files distributed with SCALE are discussed in the SCALE Nuclear Data Covariance Library chapter, and the format of the COVERX data file is presented in COVERX format. A prerequisite in the uncertainty analysis approach is that cross section uncertainty data for all nuclides and reactions of interest have been evaluated and processed for use by these procedures. However, evaluated cross section uncertainty data are not available for all nuclide-reaction pairs. Nuclide-reaction pairs without data are omitted from this analysis, but it is assumed that either the cross section data values from these pairs are well known (i.e., small uncertainties), or that the sensitivity of the system keff to these nuclide-reaction pairs is small. Where these assumptions hold, the nuclide-reaction pairs without cross section-uncertainty data present a negligible contribution to the uncertainty-based analysis. For situations where this negligible contribution assumption is judged to be invalid, the use of uncertainty analysis is not appropriate. However, the COVARIANCE data block can be used to input uncertainty values for the cross section data for particular nuclide-reaction pairs to assess the impact of additional covariance data. To utilize the covariance data generated by user input in the COVARIANCE data block, the keyword use_icov must be entered in the PARAMETER data block. Additionally, default uncertainty values can be assigned for all unknown covariance data. This default uncertainty data is input in the PARAMETER data block and the keyword use_dcov must be entered to activate its use. Warning messages are printed to identify substituted covariance matrices.
When use_dcov and/or use_icov and cov_fix are specified in the PARAMETER data block, and a reaction has zero or large (standard deviation > 1000%) values on the diagonal of the covariance matrix, these values are replaced with the square of the user input or default standard deviation values, and the corresponding off-diagonal terms are substituted according to the user input or default correlation values. Warning messages are printed to identify which values were replaced and which standard deviation value was used in the replacement.
The mathematical development of the integral index ck is presented here based on the development given in [refTsip1]. The nuclear data parameters (i.e., group wise nuclide-reaction specific cross sections) are represented by the vector \(\alpha \equiv \left(\alpha_m \right), m = 1, 2, \dots, M\), where M is the number of nuclide-reaction pairs \(\times\) the number of energy groups. The corresponding symmetric M \(\times\) M matrix containing the relative variances (diagonal elements) and covariances (off-diagonal elements) in the nuclear data are
where
where \(\delta \alpha_m\) and \(\delta \alpha_p\) represent the difference between the values and expectation values of the nuclear data parameters and represents integration over the ranges of \(\alpha_m\) and \(\alpha_p\) weighted with a probability density function. A rigorous definition of the cross section covariance data are given in [refTsip5].
The matrix containing sensitivities of the calculated keff to the \(\alpha\) parameters is given as
where I is the number of critical systems being considered. In TSUNAMI-IP, the elements of \(S_k\) are the sensitivity coefficients read from the sensitivity data files identified in the APPLICATIONS and EXPERIMENTS data blocks. The uncertainty matrix for the system keff values, \(C_{\bf{\text{kk}}}\), is given as
where \(\dagger\) indicates a transpose.
\(S_k\) is an I \(\times\) M matrix; \(C_{\bf{\alpha \alpha}}\) is an M \(\times\) M matrix; and the resulting \(C_{\bf{\text{kk}}}\) matrix is of dimension I \(\times\) I. The \(C_{\bf{\text{kk}}}\) matrix consists of relative variance values, \(\sigma _{i}^{2}\), for each of the systems under consideration (the diagonal elements), as well as the relative covariance between systems, \(\sigma_{ij}^{2}\) (the off-diagonal elements). These off-diagonal elements represent the shared or common variance between two systems. The off diagonal elements are typically divided by the square root of the corresponding diagonal elements (i.e., the respective standard deviations) to generate a correlation coefficient matrix. Thus, the correlation coefficient is defined as
such that the single \(c_k\) value represents the correlation coefficient between uncertainties in system i and system j.
These correlations are primarily due to the fact that the uncertainties in the calculated keff values for two different systems are related, since they contain the same materials. Cross section uncertainties propagate to all systems containing these materials. Systems with the same materials and similar spectra would be correlated, while systems with different materials or spectra would not be correlated. The interpretation of the correlation coefficient is the following: a value of 0.0 represents no correlation between the systems, a value of 1.0 represents full correlation between the systems, and a value of -1.0 represents a full anti-correlation.
To request the computation of \(c_k\) for each application identified in the APPLICATIONS section of the input in relation to each experiment in the EXPERIMENTS portion of the input, simply enter the input c in the PARAMETER section of the input. Additionally, the values table must be requested to output the full listing of \(c_k\) values. The csummary edit prints a listing of all experiments that have a \(c_k\) value exceeding the criteria value set by cvalue=.
6.3.1.1.2. Integral index c\(_r\)
The integral index \(c_k\) is intended for investigative analysis and is defined as \(c_k\) with user-defined reactions removed from consideration. The EXCLUSIONS data block is used to identify which reactions will be excluded from consideration in the calculation of \(c_k\). If the user identifies no reactions, then \(c_r\) will compute the same value as \(c_k\). Using \(c_r\) is equivalent to removing all sensitivities for a given reaction or series of reactions for all nuclides from the sensitivity data file for all applications and all experiments considered in the analysis.
To request the computation of \(c_r\) for each application identified in the APPLICATIONS section of the input in relation to each experiment in the EXPERIMENTS portion of the input, simply enter the input cr in the PARAMETER section of the input. Additionally, the values table must be requested to output the full listing of \(c_r\) values. The crsummary edit prints a listing of all experiments that have a \(c_r\) value exceeding the criteria value set by crvalue=.
6.3.1.1.3. Integral index E
The E index is a replacement for the previous Esum index, which was present in TSUNAMI-IP for SCALE 5.0 and 5.1.1. If the group-wise sensitivity data for all nuclides and reactions for each system is thought of as a vector, then the integral index E is the cosine of the angle between the two sensitivity vectors for the analyzed systems. If the two sensitivity vectors are parallel, i.e., proportional, the systems are similar. E does not require cross section covariance data and is normalized such that an E value of 0.0 indicates the systems are totally dissimilar, and an E value of 1.0 indicates the two systems are the same. Mathematically, an E value as low as -1.0 could be generated, but this would be the result of a rare combination of system sensitivity coefficients such that the sensitivity of the respective system responses would have to be exactly proportional in magnitude and opposite in sign, which seems not to be physically feasible. The E parameter is considered global in nature because its single quantity assesses similarity between two systems based on the magnitude and shape of all sensitivity profiles. The vector \(\mathbf{S}_{\mathbf{i}}\) is defined as the sensitivity vector (not matrix) for a particular application or experiment “i.” The magnitude of the sensitivity vector corresponds to the L2 norm: \(\left|\mathbf{S}_{\mathbf{i}}\right|=\sqrt{\mathbf{S}_{\mathbf{i}}^{\mathbf{T}} \mathbf{S}_{\mathbf{i}}}\). The E value for a given application a with experiment e is then
The similarity of systems in terms of their sensitivities to only the fission, capture or scatter reactions can also be evaluated as
where the vectors \(S_{x,a}^{{}}\) and \(S_{x,e}^{{}}\) represent the sensitivity of application and experiment to fission, capture, or scatter reaction x.
To request the computation of E for each application identified in the APPLICATIONS section of the input in relation to each experiment in the EXPERIMENTS portion of the input, simply enter the input e in the PARAMETER section of the input. Additionally, the values table must be requested to output the full listing of E values. The reaction components of E (i.e., Ef, Ec and Es) are included in the values table if prtparts is entered in the input. The reaction-specific components of E are each normalized between -1 and 1, the same as E itself. The esummary edit prints a listing of all experiments that have an E value exceeding the criteria value set by evalue=.
6.3.1.1.4. Integral index G
The G index assesses the similarity of two systems based on normalized differences in the energy-dependent sensitivity data for fission, capture, and scatter [refTsip2]. The similarity measure used for G is based on the concept of coverage of the application by an experiment. A physical interpretation of the G index is the ratio of the sum of the sensitivity coefficients of the application that are covered by the experiment to the sum of the sensitivity coefficients for the application. The G index, sometimes referred to as “big G” is defined as:
where,
(6.3.9)\[\begin{split}S_{x,j}^{{e}',n}=\left\{ \begin{array}{*{35}{l}} S_{x,j}^{e,n}\text{, where} & \left| S_{x,j}^{a,n} \right|\ge \left| S_{x,j}^{e,n} \right|\text{ and }\frac{S_{x,j}^{a,n}}{\left| S_{x,j}^{a,n} \right|}=\frac{S_{x,j}^{e,n}}{\left| S_{x,j}^{e,n} \right|} \\ S_{x,j}^{a,n}\text{, where} & \left| S_{x,j}^{a,n} \right|<\left| S_{x,j}^{e,n} \right|\text{ and }\frac{S_{x,j}^{a,n}}{\left| S_{x,j}^{a,n} \right|}=\frac{S_{x,j}^{e,n}}{\left| S_{x,j}^{e,n} \right|} \\ \text{0,} & \text{otherwise} \\ \end{array} \right.\end{split}\]
the n summation is performed over all nuclides present in the application system,
the x summation is performed over fission, capture and scatter reactions (f, c, and s) as appropriate for each nuclide, and
the j summation is performed over all energy groups.
The use of 1 minus the normalized difference makes the range of this index consistent with ck and Esum. Hence, a G of 1 indicates complete similarity and a G value of 0 indicates no similarity.
The definition of \(S_{x,j}^{{e}',n}\) restricts the coverage of the application by the experiment to the portion of the experiment’s sensitivity coefficient that does not exceed that of the application in magnitude. Additionally, the application’s sensitivity coefficient and that of the experiment must have the same sign. The coverage for 1H scatter for an example application and experiment is illustrated in Fig. 6.3.1 where the energy-dependent sensitivity profiles for the application, the experiment and the coverage of the application by the experiment are shown in green, red and blue, respectively. Because the sensitivity coefficients for the application and the experiment have opposite signs at energies just above 1 \(\times\) 10-2 eV, the application provides no coverage for the experiment for these groups. Also, for several groups, the sensitivity of the application exceeds that of the experiment and only partial coverage is provided. Partial coverage is illustrated where the application data (green) exceeds the coverage (blue). In other groups, the sensitivity of the experiment exceeds that of the application. In these groups, full coverage is provided, but the coverage is not allowed to exceed the sensitivity of the application. This is illustrated in groups where the experiment data (red) exceeds the coverage data (blue). With the limitation of the coverage as the value of the applications sensitivity coefficient, the experiment cannot provide “extra credit” in coverage for sensitivity coefficients that exceed those of the application.
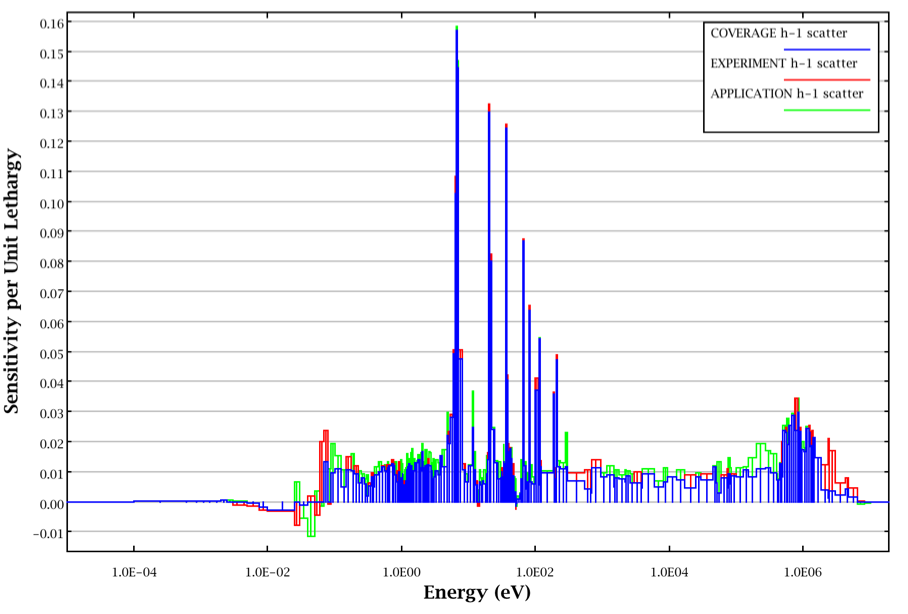
Fig. 6.3.1 Illustration of coverage for 1H scatter.
The coverage for 10B capture for an example application and experiment is illustrated in Fig. 6.3.2. In this figure, the sensitivity coefficients are all negative. The magnitudes of the sensitivity coefficients for the experiment far exceed those of the application at thermal energies. However, coverage is only provided to the magnitudes of the sensitivity coefficients of the application.
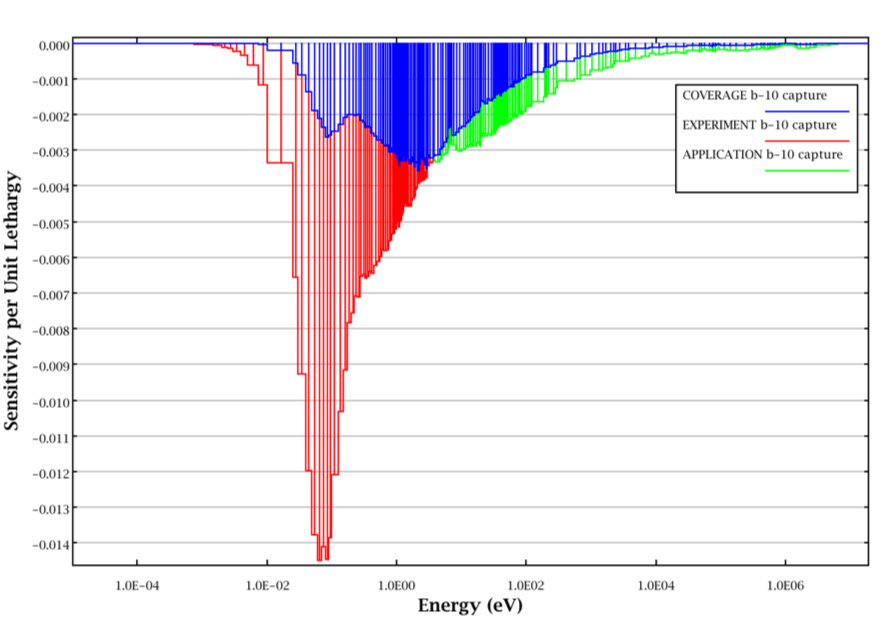
Fig. 6.3.2 Illustration of coverage for 10B capture.
Assessment of similarity over a particular reaction type (fission, capture or scatter) can be made with a partial G value. A \(G_x\) value can be computed by eliminating the sum over reactions, x in Eq. (6.3.11), as
where
x is f, c, or s.
To request the computation of G for each application identified in the APPLICATIONS section of the input in relation to each experiment in the EXPERIMENTS portion of the input, simply enter the input g in the PARAMETER section of the input. Additionally, the values table must be requested to output the full listing of G values. The reaction components of G (i.e., \(G_f\), \(G_c\) and \(G_s\)) are included in the values table if prtparts is entered in the input. The \(G_x\) indices are normalized such they also have a range of 0 to 1, where a \(G_x\) of 1 indicates complete similarity of sensitivity coefficients for that particular reaction type x between the application and experiment. The gsummary edit prints a listing of all experiments that have a G value exceeding the criteria value set by gvalue=.
6.3.1.2. Nuclide-reaction specific integral indices
The global integral indices described in Sect. 6.3.1.1 assess system similarity for all nuclides and reactions in the application system. It is also possible and sometimes desirable to produce values analogous to G, E, and ck for each nuclide-reaction pair, such that similarity can be assessed on a nuclide-reaction-specific level.
6.3.1.2.1. Nuclide-reaction specific integral index g
The nuclide-reaction specific integral index based on the same coverage criteria as G is denoted g, and sometimes referred to as “little g.” [2] It is defined in terms of the normalized differences of the group wise sensitivity coefficients for a particular nuclide, n, and reaction, x, summed over all energy groups, j, as
where,
(6.3.12)\[\begin{split}S_{x,j}^{{e}',n}=\left\{ \begin{array}{*{35}{l}} S_{x,j}^{e,n}\text{, where} & \left| S_{x,j}^{a,n} \right|\ge \left| S_{x,j}^{e,n} \right|\text{ and }\frac{S_{x,j}^{a,n}}{\left| S_{x,j}^{a,n} \right|}=\frac{S_{x,j}^{e,n}}{\left| S_{x,j}^{e,n} \right|} \\ S_{x,j}^{a,n}\text{, where} & \left| S_{x,j}^{a,n} \right|<\left| S_{x,j}^{e,n} \right|\text{ and }\frac{S_{x,j}^{a,n}}{\left| S_{x,j}^{a,n} \right|}=\frac{S_{x,j}^{e,n}}{\left| S_{x,j}^{e,n} \right|} \\ \text{0,} & \text{otherwise} \\ \end{array} \right.\end{split}\]and the j summation is performed over all energy groups.
The criteria for g are the same as those explained in Sect. 6.3.1.1.3. The only difference in g and G is that the summations over nuclide and reaction have been removed. As with other integral indices, the g index is normalized such that a g value of 1 indicates complete coverage of the application by the experiment for the particular nuclide-reaction pair. A g value of 0 indicates no coverage of the application by the experiment for the particular nuclide-reaction pair.
To request the computation of g (“little g”) for each application identified in the APPLICATIONS section of the input in relation to each experiment in the EXPERIMENTS portion of the input for fission, capture and scatter reactions for each nuclide in the application, simply enter the input lg in the PARAMETER section of the input. The lg data appears in the nuclide-reaction specific edit for each application and only includes nuclide-reaction pairs with energy integrated sensitivity coefficients with at least the magnitude of sencut. Additionally, g values are printed for each nuclide-reaction pair specified in the REACTIONS data block. The lgall edit prints all the g values for fission, capture and scatter for all nuclides in the application in relation to all experiments regardless of the magnitude of the energy-integrated sensitivity value. The lgsum edit prints a listing of all g values exceeding the criteria value set by lgvalue= for fission, capture and scatter reactions for each nuclide. The lggroups edit prints a long table listing the numbers of experiments that are at least as sensitive as the application for each group for each nuclide-reaction pair requested in the REACTIONS data block.
6.3.1.2.2. Extended \(c_k\)
The extended \(c_k\) edit lists the contribution of each nuclide-reaction-to-nuclide-reaction energy covariance matrix to the global integral index \(c_k\). A single entry in this edit is computed by utilizing a subset of the \(\mathbf{C}_{\mathbf{\alpha \alpha}}\) matrix and the \(\mathbf{S}_{\mathbf{k}}\) sensitivity vectors for the application and the experiment. Thus, the extended \(c_k\) for a nuclide-reaction pair in the application in relation to a nuclide-reaction pair in the experiment is defined as
where,
(n-j)a represents the nuclide n and reaction j of the application a, (m-k)e represents the nuclide m and reaction k of the experiment e, \(\sigma _{{{\left( n-j \right)}_{a}},{{\left( m-k \right)}_{e}}}^{2}\) represents the covariance between application a and experiment e due to this nuclide-reaction pairs, \(\sigma_a\) is the standard deviation in keff for the application due to all cross section covariance data, and \(\sigma_e\) is the standard deviation in keff for the experiment due to all cross section covariance data.
The global integral index \(c_k\) can be reconstructed from the extended \(c_k\) components by summing the extended \(c_k\) over all nuclide-reaction-to-nuclide-reaction values. Thus, the extended \(c_k\) allows the user to determine the contribution of nuclide-reaction-to-nuclide-reaction covariances to \(c_k\). All covariance data, default covariance data and user-input covariance data used in the calculation of \(c_k\), as outlined in Sect. 6.3.1.1, are also applied to the extended \(c_k\) data. Nuclide-reaction-to-nuclide-reaction covariances using default and user-input values are identified with one and two asterisks, respectively, in the text output, and are identified with unique colors in the HTML output. Nuclide-reaction-to-nuclide-reaction covariances using default and user-input values for cov_fix adjustments are identified with three and four asterisks, respectively, in the text output, and are identified with unique colors in the HTML output. Warning messages are printed to identify which zero values were replaced and which standard deviation value was used in the substitution.
The extended \(c_k\) edit also includes an individual \(c_k\) to examine the similarity of an application and experiment based only on a single nuclide-reaction pair. The individual \(c_k\) is similar to the \(c_k\) contribution from Eq. (6.3.13), except that it is normalized between -1 and 1 for each nuclide-reaction pair as
Note that individual \(c_k\) values computed for a nuclide-reaction pair in an application to the same nuclide-reaction pair in the experiment. Although cross-reaction and cross-nuclide covariance data are available, the cross-relationship has no physical interpretation for assessing for assessing the similarity of systems for a specific nuclide-reaction pair.
To request the computation of extended \(c_k\) for each application identified in the APPLICATIONS section of the input in relation to each experiment in the EXPERIMENTS portion of the input, simply enter the input c_long in the PARAMETER section of the input.
6.3.1.2.3. Extended c\(_r\)
Similar to the extended \(c_k\) edit, the extended \(c_r\) edit lists the contribution of each nuclide-reaction-to-nuclide-reaction energy covariance matrix to the global integral index \(c_k\). An individual \(c_r\) evaluating the similarity of each application to each experiment in terms of only one nuclide-reaction pair, normalized from -1 to 1, is also available. The computation of the extended \(c_r\) is the same as that of extended \(c_k\), but with user-defined reactions removed from consideration. The EXCLUSIONS data block is used to identify which reactions will be excluded from consideration in the calculation of \(c_r\) and its components as presented in extended \(c_r\). If the user identifies no reactions, then \(c_r\) and extended \(c_r\) will be computed the same as \(c_k\) and extended \(c_k\). The \(c_r\) parameter and extended \(c_r\) edit are equivalent to removing all sensitivities for a given reaction or series of reactions from the sensitivity data file for all applications and all experiments considered in the analysis.
As with extended \(c_k\), nuclide-reaction-to-nuclide-reaction covariances using default and user-input values are identified with one and two asterisks, respectively, in the text output, and are identified with unique colors in the HTML output. Nuclide-reaction-to-nuclide-reaction covariances using default and user-input values for cov_fix adjustments are identified with three and four asterisks, respectively, in the text output, and are identified with unique colors in the HTML output. Warning messages are printed to identify which values were replaced and which standard deviation value was used in the substitution.
To request the computation of extended \(c_r\) for each application identified in the APPLICATIONS section of the input in relation to each experiment in the EXPERIMENTS portion of the input, simply enter the input cr_long in the PARAMETER section of the input.
6.3.1.3. Penalty assessment
A method is available to assess an additional margin to subcriticality, or penalty, where sufficient experiments are not available to provide complete coverage for a particular application [refTsip3]. This penalty is intended as an additional uncertainty component that can be added to the calculated value of keff to provide an added measure of safety for application systems where validation coverage is lacking. The penalty calculation is based on the criteria for coverage explained in Sect. 6.3.1.1.3. The criteria for coverage in this implementation of the penalty assessment is that if a single experiment that passes qualification tests for the particular application exhibits a sensitivity coefficient for a particular energy group for a particular nuclide-reaction pair that is at least as great in magnitude and has the same sign as the corresponding sensitivity coefficient for the application, then adequate coverage exists for the code validation of the application. For group-wise nuclide-reaction specific sensitivity coefficients for the application that are not fully covered by the experiments, the uncovered portion of the sensitivity coefficient is used to compute an uncertainty in keff though the cross section covariance data.
Any experiment used in the penalty assessment calculation must pass a qualification test to determine global similarity of the experiment, based on a global integral index (\(c_k\), E, or G), and may also have to pass a nuclide-reaction specific qualification test based on the g integral index. Thus, only experiments that exhibit a certain degree of similarity to the application can be considered in this calculation, unless the tests are deactivated at the user’s discretion. Additionally, a number of similar experiments may be required before any penalty assessment is produced.
To compute the penalty, a vector of the minimum differences in the sensitivity coefficients, \(\mathbf{Z}_{\mathbf{a}}\), for the application with respect to all experiments can be obtained as
where,
\(Z_{x,j}^{a,n}=S_{x,j}^{a,n}-C_{x,j}^{a,n}\),
\(C_{x,j}^{a,n}\) is a composite of the best available sensitivity data from all experiments and is defined as
\(C_{x,j}^{a,n} = S_{x,j}^{{e}',n}\) for the experiment that satisfies \(\text{min}\left| S_{x,j}^{a,n}-S_{x,j}^{{e}',n} \right|\), e\(^{\prime}\) =1,…,E,
a represents a particular application,
e represents a particular experiment,
n represents a particular nuclide,
x represents a particular reaction,
j represents a particular energy group,
N = number of nuclides in the application system,
X = number of reactions for each nuclide,
J = number of energy groups, and
E = number of experiments meeting the qualification tests.
Once \(\mathbf{Z}_{\mathbf{a}}\) is computed, the portion of the sensitivity of the application that is not covered by the experiments can be used to propagate the uncertainty in the cross section data to a relative uncertainty in keff as
where \(\dagger\) indicates a transpose and \(\mathbf{C}_{\mathbf{\alpha \alpha}}\) is the matrix of the cross section covariance data defined in Eq. (6.3.1). In the above equation, the elements of \(Z_{\mathbf{a}}\) are each expressed in terms of \((\Delta {{k}_{\mathrm{eff}}}/{{k}_{\mathrm{eff}}})/(\Delta \Sigma /\Sigma )\), and the elements of \({{C}_{\mathbf{\alpha \alpha}}}\) are expressed in terms of relative variances or covariances as \({{(\Delta \Sigma /\Sigma )}^{2}}\), so that the final penalty is expressed as a relative uncertainty in keff , \(\Delta {{k}_{\mathrm{eff}}}/{{k}_{\mathrm{eff}}}\). This relative uncertainty in keff is written to the output file in the penalty calculations.
To request the penalty computation for each application identified in the APPLICATIONS section of the input in relation to all experiments in the EXPERIMENTS section of the input, simply enter the input penalty in the PARAMETER section of the input. Additionally, a list of the contribution each nuclide-reaction-to-nuclide-reaction covariance matrix to the total penalty can be viewed by entering penlong in the PARAMETER input. This creates an edit similar to the Uncertainty Information edit of the SAMS module. Each value shown in this output edit represents the relative penalty in percent (% \(\Delta k / k\)) due to the specified nuclide-reaction-to-nuclide-reaction covariance matrix. The values are sorted in order of descending magnitude. The cumulative penalty can be constructed by squaring the individual values, then adding those that had positive signs and subtracting those that had negative signs, then taking the square root. Negative values in the table result from covariance matrices that have anticorrelated values.
The qualification test for including or excluding experiments from the development of the penalty for a particular application is set by several input parameters. The purpose of the qualification test is to ensure that some relevant data are used in the calculation. Otherwise, in the limit that no relevant data are used in the penalty calculation, the \(Z_a\) minimum difference sensitivity vector becomes \(S_a\), the sensitivity vector for the application. In this case, the maximum penalty assessed is simply the uncertainty in the application’s criticality calculation due to cross section covariance data.
A global qualification test, to test the similarity of a particular experiment to the given application based on a global integral index, is configured with the keywords: penusec, penusee and penuseg. These keywords are used to produce a penalty calculation that only includes experiments with \(c_k\), E, or G values at least as great as cvalue, evalue or gvalue, respectively. The default value is penusec. If more than one of the keywords penusec, penusee or penuseg are included in the input, only the last one entered will be used. The penalty is only computed for a given application if the number of experiments passing the global integral index qualification test is at least as great as penminx (default = 10). If no global test is desired, enter penminx=0.
The pencut= keyword allows the user to set a discriminator for excluding nuclide-reaction pairs from the application with small sensitivity coefficients from the penalty calculation. If the sum of the absolute values of the group-wise sensitivity coefficients is below pencut, the nuclide reaction pair is excluded from the penalty calculation. The default value is 0.0, which includes all nuclide-reaction pairs in the penalty calculation. Additionally, penlgv= sets a discriminator that only includes experiments with a g value relative to the application for a given nuclide-reaction pair that meets or exceeds penlgv. Thus, using penlgv may allow some nuclide-reaction pairs from a given experiment to be included in the penalty calculation, but exclude others from the same experiment that do not meet the criteria. The default value is 0.0, which means include all nuclide-reaction pairs that pass the other qualification tests.
The keyword penwarn activates a penalty warning edit that details which experiments were excluded from the penalty calculation based on failing the global qualification test, and which nuclide-reaction pairs were excluded from the penalty calculation based on failing the nuclide-reaction specific tests.
The composite of the best available sensitivity data from all experiments that meet the requested criteria, as used in the calculation of the penalty, can be viewed with the composite sensitivity data for nuclides and reactions requested in the REACTIONS input data block with the input keyword prtcomp described in Sect. 6.3.1.4.3.
The \(\mathbf{C}_{\mathbf{\alpha \alpha}}\) matrix in Eq. (6.3.17) is the cross section covariance data read in the COVERX formatted data file identified by coverx=. As in the calculation of \(c_k\), nuclide-reaction pairs without available covariance data are omitted from this analysis, but it is assumed that either the cross section data values from these pairs are well known (i.e., small uncertainties), or that the sensitivity of the system keff to these nuclide-reaction pairs is small. Where these assumptions hold, the nuclide-reaction pairs without cross section uncertainty data present a negligible contribution to the penalty calculation. For situations where this negligible contribution assumption is judged not to be valid, the use of uncertainty analysis for the computation of a penalty is not appropriate. However, the COVARIANCE data block can be used to input uncertainty values for the cross section data for particular nuclide-reaction pairs to assess the impact of additional covariance data. To utilize the covariance data generated by user input in the COVARIANCE data block, the keyword use_icov must be entered in the PARAMETER data block. Additionally, default uncertainty values can be assigned for all unknown covariance data. The default uncertainty data input in the PARAMETER data block and the keyword use_dcov must be entered to activate its use. In the extended penalty edit, nuclide-reaction-to-nuclide-reaction covariances using default and user-input values are identified with one and two asterisks, respectively, in the text output, and are identified with unique colors in the HTML output. Warning messages are also printed to identify substituted covariance matrices.
When use_dcov and/or use_icov and cov_fix are specified in the PARAMETER data block, and a reaction has zero or large (standard deviation > 1000%) values on the diagonal of the covariance matrix, these values are replaced with the square of the user input or default standard deviation values, and the corresponding off-diagonal terms are substituted according to the user input or default correlation values. In the extended penalty edit, nuclide-reaction-to-nuclide-reaction covariances using default and user-input values for cov_fix adjustments are identified with three and four asterisks, respectively, in the text output, and are identified with unique colors in the HTML output. Warning messages are printed to identify which values were replaced and which standard deviation value was used in the substitution.
6.3.1.4. Other parameters
TSUNAMI-IP can produce a number of other parameters that are useful for analysis of systems. These are briefly explained in this section.
6.3.1.4.1. Uncertainty information
The keyword uncert activates the calculation of the uncertainty in keff due to the cross section covariance data read from the COVERX formatted data file identified by coverx=. The uncertainty is computed for each application and each experiment and is printed in the values table as a standard deviation value with its statistical uncertainty, if appropriate. The uncertainty value printed is a relative uncertainty in percent (i.e., \(\Delta {{k}_{\mathrm{eff}}}/{{k}_{\mathrm{eff}}}\) \(\times\) 100%).
Nuclide-reaction pairs without available data are omitted from this analysis, but it is assumed that either the cross section data values from these pairs are well known (i.e., small uncertainties), or that the sensitivity of the system keff to these nuclide-reaction pairs is small. Where these assumptions hold, the nuclide-reaction pairs without cross section uncertainty data present a negligible contribution to the uncertainty-based analysis. For situations where this negligible contribution assumption is judged not to be valid, the use of uncertainty analysis is not appropriate. However, the COVARIANCE data block can be used to input uncertainty values for the cross section data for particular nuclide-reaction pairs to assess the impact of additional covariance data. To utilize the covariance data generated by user input in the COVARIANCE data block, the keyword use_icov must be entered in the PARAMETER data block. Additionally, default uncertainty data can be assigned for all unknown covariance data. This default data is input in the PARAMETER data block and the keyword use_dcov must be entered to activate its use.
To request a listing of the contributions nuclide-reaction-to-nuclide-reaction covariance matrix to the uncertainty in the keff value for each application identified in the APPLICATIONS section of the input enter the input keyword uncert_long in the PARAMETER section of the input. This creates an edit similar to the Uncertainty Information edit of the SAMS module. Each value shown in this output edit represents the relative uncertainty in percent (%\(\Delta k/k\)) due to the specified nuclide-reaction-to-nuclide-reaction covariance matrix. The values are sorted in order of descending magnitude. The cumulative uncertainty can be constructed by squaring the individual values, then adding those that had positive signs and subtracting those that had negative signs, then taking the square root. Negative values in the table result from covariance matrices that have anticorrelated values.
In the extended uncertainty edit, nuclide-reaction-to-nuclide-reaction covariances using default and user-input values are identified with one and two asterisks, respectively, in the text output, and are identified with unique colors in the HTML output. Nuclide-reaction-to-nuclide-reaction covariances using default and user-input values for cov_fix adjustments are identified with three and four asterisks, respectively, in the text output, and are identified with unique colors in the HTML output. Warning messages are printed to identify which values were replaced and which standard deviation value was used in the substitution.
6.3.1.4.2. Completeness parameter
A parameter has been developed to assess the completeness of a set of experiments for the code validation of a given application. [4] The set of experiments is “complete” in the sense that it completely tests all the important cross section elements in the particular application of interest. The availability of sensitivity coefficients provides a key element in the definition of this completeness parameter.
The completeness parameter, R, is defined as follows:
where
and
\(N_{x,j}^{n}\) = number of systems for which \(S_{x,j}^{e,n}\) > | senfac \(\times\) \(S_{x,j}^{a,n}\)|
e = experiment,
a = application,
\(S_{x,j}^{e,n}\) = the sensitivity of keff of an experiment to the cross sections of the constituent material nuclide n, reaction x, and energy group j,
\(S_{x,j}^{a,n}\) = the sensitivity of keff of the application to the cross sections of the constituent material nuclide n, reaction x, and energy group j,
nixlim = an integer, and
senfac = a real number such that \(0.0 \leq\) senfac \(\leq 1.0\).
The completeness parameter is designed to give the effective fraction of the total sensitivity for each application system that is “covered” by the benchmark set. This coverage is defined by comparing the magnitude of each group-wise sensitivity coefficient for the application with respect to each of the corresponding sensitivities of the benchmark systems. The completeness parameter is computed for each application for each experiment if the keyword cp is entered in the PARAMETER data. The minimum coverage of the sensitivity coefficients for the experiment systems is defined as senfac\(\times\) 100% of the application sensitivity in the definition of \(N_{x,j}^{n}\). The value of senfac is set with the keyword input senfac=, and the default value is 0.9. Thus, the experiment must have a sensitivity coefficient at least 90% as great as that of the application for the particular nuclide, reaction and energy group to count in \(N_{x,j}^{n}\). The number of experiments counted in \(N_{x,j}^{n}\) must be at least nixlim to change d from 0 to 1, which indicates coverage for the particular nuclide, reaction and group of the application. The value of nixlim is set by the nixlim= keyword, and it has a default value of 10
6.3.1.4.3. Composite sensitivity data
A composite of the best available sensitivity data from all experiments included in the analysis, based on the coverage criteria used for the G integral index in Sect. 6.3.1.1.4 is produced for each application for each nuclide-reaction pair in the REACTIONS input if the keyword prtcomp is entered. The composite profile for a particular application a for nuclide n and reaction x is defined as the vector
where,
\(C_{x,j}^{a,n}\) is \(S_{x,j}^{{e}',n}\) for the experiment that satisfies \(\text{min}\left| S_{x,j}^{a,n}-S_{x,j}^{{e}',n} \right|\) \(e^{\prime}\) = 1, …, E,
a represents a particular application,
e represents a particular experiment,
n represents a particular nuclide,
x represents a particular reaction,
j represents a particular energy group,
J = number of energy groups, and
E = number of experiments.
An example composite sensitivity profile for 1H total is shown in Fig. 6.3.3. Here, the composite sensitivity profile comprised of the best available data from unidentified experiments 1–5 is shown in black. The sensitivity of the unidentified application is shown in red. The areas where the red application curve exceeds the black composite curve are considered uncovered. Areas where the experiment data exceed the application data are considered fully covered. Note that the composite data does not exceed the application data.
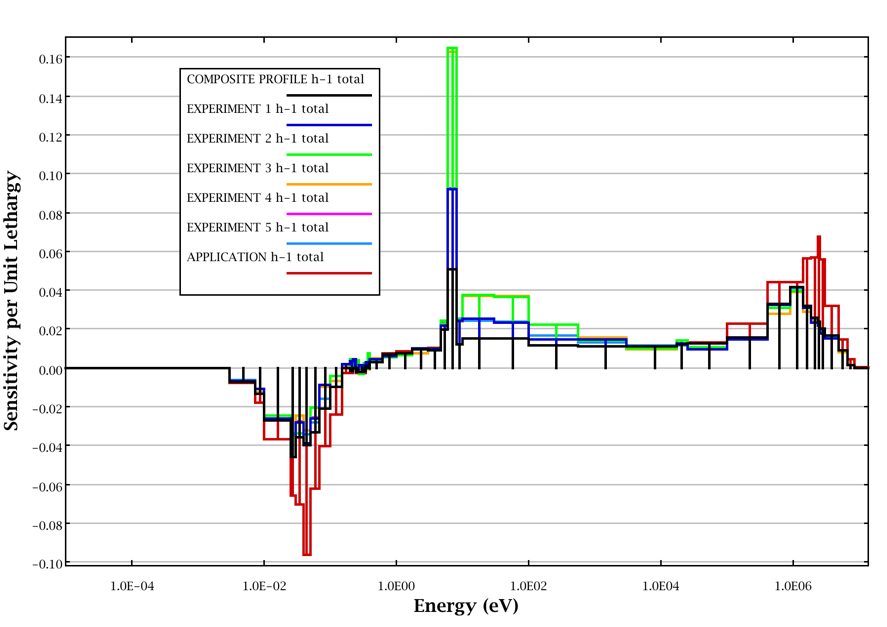
Fig. 6.3.3 Example composite sensitivity profile.
If the composite data for a particular nuclide-reaction pair for a particular energy group are added to the minimum difference data as defined in Eq. (6.3.15), the sensitivity of the application would result as
A sensitivity data file containing the composite sensitivity data is generated to permit further analysis. Only data for those nuclides and reaction identified in the REACTIONS input block are included in the composite sensitivity data file. The data file is identified with the TSUNAMI-IP input file name with the extension “.sdf” and is suitable for use with Fulcrum. This data file is presented as an interactive plot in the HTML output. The following data are included on the composite sensitivity data file for each nuclide-reaction pair identified in the REACTIONS input block for each application: the sensitivity of the application identified with “Application” and the application number, the composite sensitivity data as defined in Eq. (6.3.21) , and the composite profile used in the definition of the minimum difference profile from the penalty calculation (Sect. 6.3.1.3), which may be reduced from the full composite by excluding experiments not meeting the cutoff criteria for the penalty calculation. The composite data as used in the penalty calculation are identified as “Cut Composite.”
6.3.1.4.4. Non-coverage
A summary of non-coverage for the nuclide-reaction pairs entered in the REACTIONS section of the input can be produced if the keyword prtnotcv is entered in the PARAMETER data. The non-coverage edit gives the number of groups that are not fully covered according to the G criteria and gives the sum of the non-covered portion of the sensitivity coefficients. It also lists the group with largest sensitivity, regardless of whether or not it is covered, and gives the sensitivity value for this group. The experiment that best covers the group with the maximum sensitivity is given and the sensitivity of this group for the best matching experiment is given.
6.3.1.5. Miscellaneous options
Several input parameters that provide a wide range of options in TSUNAMI-IP are explained here. These keywords are entered in the PARAMETER data block.
6.3.1.5.1. VADER input files
VADER (formerly USLSTATS) is a data regression tool described in Ref. [refTsip6] and in the VADER chapter of the SCALE manual. The uslstats keyword requests input files for VADER to be generated from all integral indices computed by TSUNAMI-IP. The uslsummary keyword requests VADER input files to be generated from all integral indices exceeding their particular acceptance criteria (cvalue, evalue, and gvalue). The uncertainties in the keff values for VADER can be adjusted with two options. The default setting is that the uncertainty for each keff value included in the VADER input file consists of the square root of the sum of the squares of the Monte Carlo uncertainty and a uniform experimental uncertainty of 0.3%. To quadratically add the uncertainty in keff due to cross section covariance data to this uncertainty, enter the keyword usl_uncert. To modify the uniform experimental uncertainty, use the usl_sigs= keyword. When a positive value is entered for usl_sigs, this value is treated as the total uncertainty for each experiment, and no other contributions, Monte Carlo or cross section, are considered. When a negative value is entered for usl_sigs, this value is treated as the experimental uncertainty for each experiment and Monte Carlo and cross section uncertainties are added as appropriate. Currently, no option exists to input a unique experimental uncertainty for each experiment.
The VADER input files contain the extension .usl and the filenames
are presented in a descriptive format as title_xxxx_p_yyyy.usl, where
title is the filename of the TSUNAMI-IP input case, xxxx is the
application number in the TSUNAMI-IP input, p is the name of the
integral index (ck for ck, e for E, and g for G), and
yyyyy is the TSUNAMI-IP execution number (typically 0001). When the
summary inputs are requested with uslsummary, the filename is of the
format title_xxxx_p_sum_yyyy.usl, where sum denotes that this
VADER input file contains only the summary of experiments that exceed
the requested criteria for the specific integral index.
6.3.1.5.2. Covariance data directory listing
The prtmtrix keyword causes a listing of the energy covariances of the COVERX formatted cross section data file, identified by coverx=, to be printed.
6.3.1.5.3. HTML output
The keyword html causes TSUNAMI-IP to create an html formatted output. This output edit is accessed by opening the root.html file, where root is the name of the user’s input file without extensions. Additional resources for the html output are placed in new directories called root.htmd and applet_resources. The root.html directory contains files needed to display data produced by the current case and the applet_resources directory contains Java applets for data plotting within the HTML interface and can be shared by multiple output files within the same directory. The root.html file and associated directories are placed in the same location as the user’s input file. The html formatted output is color coded and more easily navigated than the standard plain text formatted output file. The html output can be customized with the HTML data block.
6.3.1.5.4. Case sensitive input
The inptcase keyword sets a flag in the SCALE free form input reader to prevent SCALE from translating all input to lower case. The purpose of this keyword is the identification of file and directory names that have upper case characters on a case-sensitive operating system. By default, the SCALE input reader translates all text to lower case. However, any input entered after inptcase is treated as case sensitive. All subsequent keyword entries and data block names must be entered in lower case, or errors will result.
6.3.1.5.5. Print filenames
The usename keyword causes TSUNAMI-IP to identify files with their file names in the code output. By default, TSUNAMI-IP identifies files according to the title on the TSUNAMI-A or TSUNAMI-B sensitivity data files. For files that have the same titles, or have long or non-descriptive titles, usename can provide for simplified interpretation of the output.
6.3.1.5.6. \(c_k\) and E differences
The keywords cechk= and cediff= allow the user to input discriminators that govern the output of warning messages when \(c_k\) and E values for a particular application and experiment differ by more than cediff and \(c_k\) and E are both at least as great as cechk.
6.3.1.5.6.1. Plots
The keyword plot causes Javapeño formatted plot (.plt) files of the global integral indices to be generated. A plot file containing all computed integral index values for each application as a function of experiment number is titled rootxxxx.plt, where root is the base name of the input file and xxxx is the TSUNAMI-IP execution number (typically 0001). Plot file containing the number of integral index values that exceeded the acceptance criteria for each application is titled rootsummaryxxxx.plt, where summary indicated it is a summary plot. Both plots are displayed in the HTML output or can be view with Fulcrum. If the .plt files are opened in Fulcrum with “Open Dataset…” each set of data can be manually added to the plot as desired by the user. If the .plt files are opened with “Open Plot…,” all data are immediately displayed.
6.3.1.5.7. Absolute sensitivity option
All global indices and parameters can also be calculated using absolute sensitivities. This capability is useful when working with TSAR-generated reactivity sensitivity data files. TSAR creates a sensitivity data file with reactivity sensitivities tabulated in either relative format or absolute format \((\Delta \rho /|\rho|) / \Delta \sigma / \sigma)\). If keyword relative is specified in the PARAMETER block, any absolute reactivity sensitivities are internally converted to \(\left(\Delta \rho\right)/\left(\Delta \sigma / \sigma \right)\). (Note that the reactivity sensitivity is normalized by \(\rho\) rather than \(|\rho\). Normalizing by \(|\rho|\) can potentially lead to \(c_k\) and E indices with the wrong sign.) Likewise, if the keyword absolute is specified, any relative reactivity sensitivities are internally converted to (\(\Delta \rho \cdot \rho / \left(\Delta \sigma / \sigma \right)\), and the SAMS-generated relative keff sensitivities are internally converted to \(\left( \Delta k_{eff} / k_{eff} \right) / \left(\Delta \sigma / \sigma \right)\). If both the relative and absolute keywords are specified, the last keyword entered in the PARAMETER block is used to determine the sensitivity format. If neither keyword is entered, the default relative format is applied.
It is important to note that the absolute sensitivity option has been added for user control over the format of the uncertainty edit and the extended uncertainty edit. Using absolute sensitivities, the absolute standard deviation in keff or \(\rho\) is edited. Using relative sensitivities, the relative standard deviation in keff or \(\rho\) is edited. The global indices \(c_k\) and E are the same regardless of the sensitivity format option. For other indices and parameters (i.e., G, penalty assessment, and completeness parameter), their respective values can change depending on sensitivity format option. These indices and parameters play an important role in nuclear criticality safety validation, and the default relative format option is recommended.
6.3.1.6. User input
The user input for TSUNAMI-IP is described in this section. The input consists of an optional title on a single line followed by one required and six optional blocks of data which are identified in Table 6.3.1 and individually described in subsequent subsections. These data blocks must begin with READ KEYNAMEand end with END KEYNAME, where KEYNAME is the name of an individual data block. The required PARAMETER data block should be entered first, followed by the remaining blocks of data in any order. Note that this is different than the SCALE 5 and SCALE 5.1 versions of TSUNAMI-IP, which allowed the blocks of data to be in any order. The SCALE 6 version will continue if the PARAMETER data block is not entered first, but the keywords absolute and use_diff_groups (explained below) will have no effect.
Keyname |
Description |
Required/Optional |
|---|---|---|
PARAMETER |
Indices and parameters to be computed are input in this section. Output edits are requested, and user-input criteria values for numeric data are entered. |
Required |
APPLICATIONS |
File paths to sensitivity data files representing application systems for which validation by the experiments are assessed are input in this section. |
Optional* |
EXPERIMENTS |
File paths to sensitivity data files representing experiments to be used in the analysis are input in this section. |
Optional* |
RESPONSE |
File paths to sensitivity data files representing experiments or applications to be used in the analysis are input in this section. |
Optional* |
REACTIONS |
Specific nuclide-reaction pairs for which analysis with certain indices and parameters are desired can be entered in this section. |
Optional |
COVARIANCE |
User input standard deviation for nuclide-reaction pairs for which cross-section-covariance data are not available can be entered in this section. |
Optional |
EXCLUSIONS |
User input reactions to exclude from \(c_r\) and Extended \(c_r\) calculations. |
Optional |
HTML |
Parameters to customize the HTML formatted output can be entered in this section. |
Optional |
*Although the EXPERIMENTS, APPLICATIONS, and RESPONSE data blocks are optional, at least one application and one experiment must be specified on the TSUNAMI-IP input file. This can be done in a variety of ways explained below.
6.3.1.6.1. Parameter data
The PARAMETER data block is used to request the calculation of the various indices and parameters available in TSUNAMI-IP, request output edits and set criteria values. The parameter block must begin with READ PARAMETER and end with END PARAMETER. The data input to the parameter data block consist of numerous keywords that are shown, along with their default values and descriptions, in Table 6.3.2. A keyword that ends with = must be followed by numeric data. Keywords that do not end with = are Boolean flags that are used to turn on certain features of the code, such as the computation of certain data or certain output edits. If the keyword is present for a Boolean entry, the value is set to true. Otherwise, the Boolean flag is set to its default value. If no data are requested in the PARAMETER section using the Boolean flags, then no data will be produced by the code. The input is designed to maximize user control over the operation of the code. A more detailed description of the indices and parameters is given in Sect. 6.3.1.1 through Sect. 6.3.1.4.
Keyword |
Default value |
Description |
absolute |
False |
Use absolute sensitivities [e.g. \(\left(\Delta k_{eff}/\Delta \sigma / \sigma\right)\)] for all applications and experiments in the analysis. If absolute is entered, the keyword relative is set to False. |
c |
False |
Compute \(c_k\) values for each application compared to each experiment. |
c_long |
False |
Produces extended \(c_k\) output edit for each application compared to each experiment. |
cechck= |
0.5 |
Level of E and \(c_k\) values that trigger the cediff warning. If E or \(c_k\) are below this value, no warning is printed. |
cediff= |
0.1 |
If the E and \(c_k\) values for a given application and experiment differ by more than cediff, a warning message is printed. |
cov_fix |
False |
Replace zero and large (standard deviation >1000%) values on diagonal of cross-section covariance data with user input values and default values. |
coverx= |
56groupcov7.1 |
Name of cross-section covariance data file to use in analysis. |
cp |
False |
Compute and print completeness parameter for each application. |
cr |
False |
Compute \(c_r\) values for each application compared to each experiment. |
cr_long |
False |
Produces extended \(c_r\) output edit for each application compared to each experiment. |
inptcase |
False |
This sets the SCALE free form reader to leave the case of the input data as read. This is useful when sensitivity data file names have upper case letters. If this option is set, all other input keywords MUST be in lower case to be correctly interpreted by SCALE. This keyword must be entered in the input prior to the reading of any titles of the sensitivity data files with capital letters. |
large_cov= |
10 |
Cutoff fractional standard deviation value for cov_fix. Covariance data with uncertainties larger than large_cov are replaced with user-defined or default values. Default = 10, which is 1000% uncertainty. |
lg |
False |
Compute g values for fission, capture and scatter for each nuclide for each experiment compared to each application and print them in a table if the application’s sensitivity for the corresponding nuclide-reaction pair is greater than or equal to sencut. Also compute g values for reactions specified in the REACTIONS data block. |
lgall |
False |
Print g values for fission, capture and scatter for all nuclides for all experiments for each application. |
lggroups |
False |
Print a table listing the numbers of experiments that are at least as sensitive as the application for each group for each reaction requested in the REACTIONS data block. |
lgsum |
False |
Print a summary table of g for each application for each experiment that exceeds lgvalue for each nuclide’s capture, fission and scatter reactions |
lgvalue= |
0.9 |
Threshold value of g for inclusion in summary table. |
nixlim= |
10 |
Minimum number of experiments with group-wise values exceeding senfac times the group-wise value for the application for the group-wise value to be added to the completeness parameter. |
penalty |
False |
Create penalty assessment based on differences in the application’s sensitivity profile for a particular nuclide-reaction pair, and the corresponding composite profile for all qualifying experiments. |
pencut= |
0.0 |
Cutoff value for excluding sensitivities from the penalty calculation. If the sum of the absolute values of the energy-dependent sensitivity data for a particular nuclide-reaction pair is below this number, the nuclide-reaction pair will not be included in the penalty calculation. |
penlgv= |
0.0 |
Use only nuclide-reaction pairs with g values exceeding penlgv in the penalty assessment. |
penlong |
False |
Print detailed edits of components of penalty assessments showing each component of the penalty. |
penminx= |
10 |
Minimum number of qualifying experiments for each application for penalty calculation. |
pensusec |
True |
Use only experiments with \(c_k\) values exceeding cvalue in the penalty assessment. If penusec is entered, the keywords penusee and penuseg are set to False. |
penusee |
False |
Use only experiments with E values exceeding evalue in the penalty assessment. If penusee is entered, the keywords penusec and penuseg are set to False. |
penuseg |
False |
Use only experiments with G values exceeding gvalue in the penalty assessment. If penuseg is entered, the keywords penusec and penusee are set to False. |
penwarn |
False |
Print list of warning messages noting the experiments and nuclide-reaction pairs that were excluded from the penalty calculation. |
plot |
False |
Produces Javapeño formatted plot (.plt) files for integral values and composite sensitivity data. |
prtcomp |
False |
Print “composite” of experiment sensitivity profiles for reactions selected in REACTIONS data block. Also write data to sensitivity data file. |
prtmtrix |
False |
Print directory of data available on the cross-section-covariance data library. |
prtnotcv |
False |
Print a table summarizing the non-coverage for the nuclide-reaction pairs entered in the REACTIONS data block. |
prtparts |
False |
Print the components of E and G for fission capture in scatter in the values table. |
relative |
True |
Use relative sensitivities [e.g., \(\left(\Delta k_{eff}k_{eff}\right)/\left(\Delta\sigma/\sigma\right)\) for all applications and experiments in the analysis. If relative is entered, the keyword absolute is set to False. |
return_work_cov |
False |
Copy the working covariance library to the return directory with the file name job_name.wrk.cov, where job_name is the name of the input file. If return_work_cov is not present, the working covariance library remains in the temporary working directory with the file name job_name.wrk. |
sencut= |
0.01 |
Cutoff value for ignoring low valued sensitivities in nuclide-reaction specific edit tables. If the absolute value of the sum of the energy-dependent sensitivity data for a particular nuclide-reaction pair is below this number, the nuclide-reaction pair will not be included in the edit. |
senfac= |
0.9 |
Value used in calculation of completeness parameter. Group-wise sensitivity for the application is counted as validated by the experiment if the sensitivity from the experiment is greater than the application sensitivity times senfac. |
udcov= |
0.05 |
User-defined default value of standard deviation in cross-section data to use for all groups for nuclide-reaction pairs for which covariance data are not available on the selected data file. |
udcov_corr= |
1.0 |
User-defined default correlation value to use for nuclide-reaction pairs for which covariance data are not available on the selected data file. |
udcov_corr_type= |
zone |
User-defined default correlation in cross-section data to use for nuclide-reaction pairs for which covariance data are not available on the selected data file. Allowed values are long, zone, and short. (See Sect. 6.3.1.6.3 for details on long, zone and short.) |
udcov_fast= |
0.0 |
User-defined default value of standard deviation in cross-section data to use for fast data for nuclide-reaction pairs for which covariance data are too large or not available on the selected data file. |
udcov_inter= |
0.0 |
User-defined default value of standard deviation in cross-section data to use for intermediate data for nuclide-reaction pairs for which covariance data are too large or not available on the selected data file. |
udcov_therm= |
0.0 |
User-defined default value of standard deviation in cross-section data to use for thermal data for nuclide-reaction pairs for which covariance data are too large or not available on the selected data file. |
uncert |
False |
Computes uncertainty in keff due to covariance data |
uncert_long |
False |
Prints extended table of uncertainty in keffdue to covariance data. |
use_dcov |
False |
Use user-defined default covariance data for nuclide reaction pairs not included on the covariance data file. The user-defined data will be used in the penalty assessment as well as the computation of ck and uncertainty calculations. |
use_diff_groups |
True |
Allow sensitivity data files to have different energy groups. Data files with different energy groups will be internally converted to the energy group structure of the covariance data file. This parameter is now always equal true and does not need to be set. |
use_icov |
False |
Use user-defined data input in COVARIANCE input data block in place of default values for user input nuclide-reaction pairs that are not on the covariance data file. The user-defined data will be used in the penalty assessment as well as the computation of ck and uncertainty calculations. |
usename |
False |
Use the name of the sensitivity data file in place of its title in all output. |
uslstats |
False |
Produces VADER input files for trending analysis with all experiments for each global integral index (ck, E, and G) requested for each application |
uslsummary |
False |
Produces VADER input files for trending analysis with experiments exceeding cutoff value (cvalue, evalue or gvalue) for each global integral index (ck, E, and G) requested for each application |
usl_p= |
0.9990 |
Value of P in VADER input files. P is the portion of the population falling above the lower tolerance level |
usl_1-g= |
0.9500 |
Value of 1-\(\gamma\) in VADER input files. 1-\(\gamma\) is the confidence on the fit. |
usl_alpha= |
0.9990 |
Value of \(\alpha\) in VADER input files. \(\alpha\) is the confidence on the proportion of P. |
usl_xmin= |
0.0000 |
Value of x(min) in VADER input files. x(min) is the minimum value of the parameter x. |
usl_xmax= |
1.0000 |
Value of x(max) in VADER input files. x(max) is the maximum value of the parameter x. |
usl_sigs= |
-0.0030 |
Value of \(\sigma\)s in VADER input files. If a positive value is entered, it is applied as the total uncertainty for each experiment. If a negative or zero value is entered, it is used as the experimental measurement uncertainty and any Monte Carlo and cross section uncertainties are added quadratically to the absolute value of usl_sigs for each experiment. |
usl_dkm= |
0.0500 |
Value of \(\Delta k_m\) in VADER input files. \(\Delta k_m\) is the administrative margin used to ensure subcriticality. |
usl_uncert |
False |
Includes uncertainty in keff due to cross section covariance data from keff uncertainty written to VADER input files. |
values |
False |
Print all computed “global” indices (E, \(c_k\), and G) in a table for each application. If prtparts is input, also include the partial values |
6.3.1.6.2. Reaction data
The REACTIONS data block is used to specify nuclide-reaction pairs for the lg and lggroups edits. The reactions block must begin with READ REACTIONS and end with END REACTIONS. Data are entered in pairs with the nuclide number (e.g., 92235) followed by the reaction MT number (e.g., 18). Alphanumeric input is also accepted (e.g., u-235 fission) for the nuclide-reaction pairs. Mixed input is also acceptable (e.g., 92235 fission). Available reaction types are given in Table 6.3.4.
6.3.1.6.3. User input covariance data
The COVARIANCE data block allows the user to specify a covariance matrix for specific nuclide-reaction pairs for which covariance data are not present on the covariance data file or that have zero or large values on the diagonal. The COVARIANCE data block must begin with READ COVARIANCE and end with END COVARIANCE.
Input Parameter |
Requirement |
Default Value |
Allowed Values |
Description |
|---|---|---|---|---|
Nuclide |
Required |
none |
Nuclide name or ZA number |
Nuclide for which covariance data are to be entered |
Reaction |
Required |
none |
Reaction name or MT number |
Reaction for which covariance data are to be entered. See Table 6.3.4 for available reaction types. |
all= |
Optional |
0.0 |
any number |
Fractional standard deviation value to be applied to all groups. |
fast= |
Optional |
0.0 |
any number |
Fractional standard deviation value to be applied to fast groups. The fast value overrides the all value in the fast groups. |
therm= |
Optional |
0.0 |
any number |
Fractional standard deviation value to be applied to thermal groups. The therm value overrides the all value in the thermal groups. |
inter= |
Optional |
0.0 |
any number |
Fractional standard deviation value to be applied to intermediate groups. The inter value overrides the all value in the intermediate groups. |
corr= |
Optional |
1.0 |
any number from -1.0 to 1.0 |
Correlation between groups (see corr_type for use) |
corr_type |
Optional |
zone |
long, short, zone |
Type of correlation applied form group-to-group covariance values. long – correlation is applied between all groups short – correlation is applied only between adjacent groups zone – correlation is applied within fast, intermediate and thermal groups, but no correlation is applied between zones |
end |
Required |
Denotes end of input for current nuclide/reaction |
Any MT number or reaction name will be treated as a valid input, but only those present on the sensitivity data files will produce useful information. The reaction sensitivity types computed by SAMS from TSUNAMI-1D and TSUNAMI-3D are shown in Table 6.3.4. An energy-covariance matrix is created for the specified nuclide-reaction pair with the square of the entered standard deviation for the diagonal terms for all groups using the all= value. Groups in the fast, intermediate and thermal energies are then set to the square of the standard deviation value entered for fast=, inter=, and therm=, respectively. The off-diagonal terms of the energy matrix are generated according to the input for corr=, and corr_type=, with default settings of 1.0 and zone. Data entered in this block do not override data present on the covariance data file. The SCALE 5.1 input format is supported where data are entered in triplets with the nuclide name (e.g., u-235), then the reaction MT number or name (e.g., 18 or fission), and then a standard deviation value. In this case, the end card must not be entered. The standard deviation value is applied to all groups with default setting for correlations. These data are only used if use_icov is specified in the PARAMETER data block.
MT |
Reaction |
SCALE identifier |
|
0 |
Sum of scattering |
scatter |
|
1 |
Total |
total |
|
2 |
Elastic scattering |
elastic |
|
4 |
Inelastic scattering |
n,n’ |
|
16 |
n,2n |
n,2n |
|
18 |
Fission |
fission |
|
101 |
Neutron disappearance |
capture |
|
102 |
n,\(\gamma\) |
n,gamma |
|
103 |
n,p |
n,p |
|
104 |
n,d |
n,d |
|
105 |
n,t |
n,t |
|
106 |
n,3He |
n,he-3 |
|
107 |
n,\(\alpha\) |
n,alpha |
|
452 |
\(\bar{\nu}\) |
nubar |
|
1018 |
\(\chi\) |
Chi |
If use_icov is specified, these data are used for the calculation of the uncertainty in keff (uncert and uncert_long edits), calculation of \(c_k\), Extended \(c_k\), and the penalty assessment for each application. These user input values are only applied where an application has a sensitivity profile for which there is no corresponding covariance matrix on the covariance data file. When both use_icov and cov_fix are specified in the PARAMETER data block, and a reaction has zero or large (standard deviation > 1000%) values on the diagonal of the covariance matrix, these values are replaced with the square of the user input standard deviation value, and the corresponding off-diagonal terms are substituted according to the values of corr and corr_type.
Specifying user input covariance data for the summative reactions total, scatter and capture have no affect on the results of TSUNAMI-IP, as the summative reactions are not used in uncertainty analysis calculations.
6.3.1.6.4. Application, experiment, and response data
Sensitivity data files are designated as either application systems or experiment systems by the APPLICATIONS, EXPERIMENTS, or RESPONSE data blocks. For each application system, TSUNAMI-IP will calculate integral parameters against each experiment system. Each data block must begin with READ KEYNAME and end with END KEYNAME where KEYNAME can be APPLICATIONS, EXPERIMENTS, or RESPONSE.
Inside each data block, sensitivity data files are listed using response definition records. A response definition record is a single line of input that contains the sensitivity data filename and nine optional keyword specifications. The sensitivity data filename and optional keywords can be entered in any order, with the following format:
filename (name=N)(use=U)(type=T) (omit) (ev=E) (uv=U) (nu=P) (absolute or abs) (relative or rel)
where
filename = sensitivity data filename. The filename can include the file path.
N = A descriptive identifier for the sensitivity data file in TSUNAMI-IP output edits. The identifier is limited to 20 alphanumeric characters (spaces are not allowed).
U = The file usage. Allowed values are appl, expt, and omit, signifying the use of the file as an application or an experiment or to exclude the file from the analysis, respectively. The default values for the use keyword are described below.
T = 8-character alphanumeric identifier for the response type (e.g. ‘keff’). The response type is used in various output edits along with the name=N identifier.
omit - Optional keyword used to omit filename from the analysis, can be used independent of use=.
E = experimental value of the response (e.g. keff).
U = uncertainty value of the response.
P = number of uncertainty components to characterize the experiment uncertainty for this response.
absolute - Optional keyword that specifies absolute sensitivities will be used for filename.
relative - Optional keyword that specifies relative sensitivities will be used for filename.
By default, filenames listed in the APPLICATIONS data block are designated as application systems. Likewise, filenames listed in the EXPERIMENTS or RESPONSE data blocks are designated as experiment systems. The sensitivity data files must be in either the TSUNAMI-A or TSUNAMI-B file format, detailed in Appendix Data File Formats. (TSUNAMI-IP is not currently compatible with, for example, sensitivity data files in the “ABBN” format.) Only the region-integrated sensitivity coefficients from the sensitivity data files are used by TSUNAMI-IP. These data are energy-dependent, but any mixture-dependent or region-dependent data present on the data files are not used. Case-sensitive filenames and their file paths are allowed. However, spaces are not allowed in the filenames or file paths.
The nine optional keywords provide more user control over how each sensitivity data file is used in the analysis, and how each sensitivity data file is identified in the TSUNAMI-IP output. By default, TSUNAMI-IP identifies files in the output according to the title on the sensitivity data files. For files that have the same titles, or have long or non-descriptive titles, the usename keyword in the PARAMETER data block can be used to identify sensitivity data files by their filename. Although file names are unique, they can also be non-descriptive. For this reason, the name keyword on the response definition record can be used to create a new identifier for the sensitivity data file in the TSUNAMI-IP output. Similarly, the type keyword can be used to identify the response type in the output. Because TSUNAMI-IP was initially intended for similarity assessment of critical systems, the default response type identifier is ‘keff’; the type identifier for Generalized Perturbation Theory reaction rate sensitivity responses is ‘gpt’. For reactivity sensitivity data files produced by the TSAR module (Chapter 6.4H), the default response type identifier is \(\rho\). The type keyword can be used to specify a descriptive response type identifier, such as type=k_sdm or type=alpha_t.
The use keyword specifies how the sensitivity data file is used in the analysis. Allowed values are expt, appl, or omit. All sensitivity data files on response definition records with use=expt are designated as experiment systems, regardless of what data block it is in. Likewise, all sensitivity data files on response definition records with use=appl are designated as application systems. In addition, the user can omit the sensitivity data file from the analysis by entering either use=omit or simply omit on the response definition record.
The experiment value of the response (ev=) and the experiment uncertainty of the response (uv=) are used in generating VADER input files for trending analysis. If the experiment value is not provided, then the default value used in VADER is 1.0. If the experiment uncertainty value is not provided, then the default used in VADER is the absolute value of the usl_sigs value in the PARAMETER data block.
The ev= and uv= keywords do not impact the computed results in TSUNAMI-IP and are only used if the uslstats keyword is included in the PARAMETER data block. Experiment values of keff or reactivity only need to be provided for experiment systems. TSUNAMI-IP skips over ev= and uv= keyword specifications given for application or omitted systems.
In addition to the uv= keyword specification, TSUNAMI-IP allows for the experiment uncertainty value to be given in terms of uncertainty components. The nu= keyword defines the number of uncertainty components that characterize the experiment uncertainty. If the experiment response uncertainty is given in terms of uncertainty components, the uv= keyword specification is optional. An uncertainty component definition record follows the response definition record if the nu= keyword specification is given. The uncertainty component definition record has the following format:
uncmp1 val1 uncmp2 val2 …….. uncmpP valP
where
uncmp1 =4-character alphanumeric identifier for the 1st uncertainty component.
val1 =experiment uncertainty for component uncmp1.
uncmp2 =4-character alphanumeric identifier for the 2nd uncertainty component.
val2 =experiment uncertainty for component uncmp2.
uncmpP =4-character alphanumeric identifier for the Pth uncertainty component.
valP =experiment uncertainty for component uncmpP.
The uncertainty component definition record contains nu=P pairs of alphanumeric identifiers and numeric values. The experiment uncertainty value is determined by taking the square root of the sum of the squares of each uncertainty component value.
The final optional keywords abs and rel are used to determine the format of sensitivity and uncertainty data on the response definition record and the uncertainty component definition record. For a keff response, the following four input definitions produce equivalent uncertainties:
1) name=exp_001 ev=1.001 uv=0.005000 rel C:\sensitivity\k_critical_a.sdf
2) name=exp_001 ev=1.001 uv=0.005005 abs C:\sensitivity\k_critical_a.sdf
3) name=exp_001 ev=1.001 nu=2 rel C:\sensitivity\k_critical_a.sdf
enri 0.003000 sden 0.004000
4) name=exp_001 ev=1.001 nu=2 abs C:\sensitivity\k_critical_a.sdf
enri 0.003003 sden 0.004004
In the example above, the measured keff is 1.001 \(\pm\) 0.5% or 1.001 \(\pm\) 0.005005. The sensitivity data filename is given as C:sensitivityk_critical_a.sdf, and the experiment response is referred to as exp_001 in the TSUNAMI-IP output. In 1), the relative format is used to specify the relative standard deviation of the measured response as 0.005. In 2), the absolute format is used to specify the absolute standard deviation of the measured response as 0.005005. Because the TSUNAMI-generated sensitivity data file is in relative format, TSUNAMI-IP internally renormalizes the relative sensitivities to absolute sensitivities. In 3), the relative format is used to specify the relative standard deviation of keff due to two components (enri and sden) as 0.003 and 0.004 respectively. Using the square root of the sum of the squares approach, the relative standard deviation of keff is computed to be 0.005. In 4), the absolute format is used to specify the absolute standard deviation of keff due to two components (enri and sden) as 0.003003 and 0.004004 respectively. The absolute standard deviation of keff is computed to be 0.005005. Like 2), the sensitivity data file is internally converted to contain absolute sensitivities.
Because of the high-degree of flexibility in designating application systems and experiment systems in the TSUNAMI-IP input, the following two methods are recommended. The first method is consistent with previous versions of TSUNAMI-IP. That is, application systems are listed in the APPLICATIONS data block and experiment systems are listed in the EXPERIMENTS data block. The second method would be to list both sets of systems in a single RESPONSE data block and use the use=U keyword specification to designate how each file is to be used. Sample input files for both methods are given for the TSUNAMI-IP example problem in Sect. 6.3.1.6.7.
6.3.1.6.5. Reactions excluded from \(c_r\)
The EXCLUSIONS data block contains lists of reaction types that will be removed from consideration in the calculation of the \(c_r\) integral index and in the extended \(c_r\) edit. Valid input for this block are reaction MT numbers or names (e.g., 18 or fission). Values must be separated by a space or a line break. Any MT number or reaction name will be treated as a valid input, but only those present on the sensitivity data files will produce useful information. The reaction sensitivity types computed by SAMS from TSUNAMI-1D and TSUNAMI-3D are shown in Table 6.3.4.
6.3.1.6.6. HTML data
The optional HTML data block is used to customize HTML formatted output. The HTML data block must begin with READ HTML and end with END HTML. The data input to the HTML data block consist of several keywords that are shown, along with their default values and descriptions, in Table 6.3.5. A keyword that ends with = must be followed by text data. For color entries, any valid html color name can be entered or the hexadecimal representation can be used if preceded by a # sign. For example, to change the background color of the html output to white, bg_clr=white and bg_clr=#ffffff have the same effect, because ffffff is the hexadecimal representation of white. An extensive list of available colors for customized output is shown in Appendix BH. Please note that not all features are supported by all browsers.
Keyword |
Default value |
Description |
bg_clr= |
papayawhip |
Background color. |
h1_clr= |
maroon |
Color used for major headings. |
h2_clr= |
navy |
Color used for sub-headings. |
txt_clr= |
black |
Color for plain text. |
lnk_clr= |
maroon |
Color for hyperlinks. |
lnk_dec= |
none |
Decoration for hyperlinks. (none, underline, overline, line-through, blink). |
vlnk_clr |
navy |
Color for visited hyperlinks. |
max_clr= |
maroon |
Color for maximum values in tables. |
cut_clr= |
navy |
Color for values in tables that exceed cutoff values. |
ud_clr= |
blue |
Color for values in tables that use default covariance data. |
ui_clr= |
Red |
Color for values in tables that use user-input covariance data. |
udfix_clr= |
royalblue |
Color for values in tables that use default corrected covariance data. |
uifix_clr= |
green |
Color for values in tables that use user-input corrected covariance data. |
6.3.1.6.7. Example TSUNAMI-IP input
An example TSUNAMI-IP input listing is given in Example 6.3.1. In this example, the optional title for this analysis is entered as “tsunami-ip example”. The parameter data are used to request that E, \(c_k\), G, and g be computed (e, c, g, and lg, respectively). The integral indices E, \(c_k\), and G will be listed in a table for each application containing the values of each index for each experiment (values). The cutoff values for E and \(c_k\) for inclusion in the summary tables are set to 0.8 (evalue=0.8 and cvalue=0.8). Summary tables listing experiments with E values exceeding evalue, \(c_k\)values exceeding cvalue will be produced (esummary, and csummary, respectively). The REACTIONS data block requests that the n,\(\gamma\) reaction for 235U and the elastic scattering reaction for 1H be assessed for coverage with the g index.
=shell
copy c:\applications\1sen.sdf
copy c:\applications\2sen.sdf
copy c:\applications\3sen.sdf
end
=tsunami-ip
tsunami-ip example
read parameter
e c g lg
values
evalue=0.8
cvalue=0.8
esummary csummary tesum tcsum
end parameter
read applications
1sen.sdf
2sen.sdf
3sen.sdf
end applications
read experiments
c:\sensitivity\bnwl2129t3-01sen.sdf
c:\sensitivity\bnwl2129t3-02sen.sdf
c:\sensitivity\bnwl2129t3-03sen.sdf
c:\sensitivity\bnwl2129t3-04sen.sdf
c:\sensitivity\bnwl2129t3-05sen.sdf
end experiments
read reactions
u-235 n,gamma
h-1 elastic
end reactions
end
The application systems are represented by the sensitivity data files named 1sen.sdf, 2sen.sdf and 3sen.sdf. In this example input, these data files are copied (or they could be linked in UNIX) to the temporary directory where SCALE executes and writes its scratch files via system commands that are input to the SHELL utility at the top of the input file.
The experiments for which the indices will be computed for each application are listed in the experiments data block. Here, an explicit file path is specified in the input. This could also have been done in the APPLICATIONS data block. In this case, the 5 data files would have to exist in a directory called c:\sensitivity. An equivalent input file using a single RESPONSE data block is given in Example 6.3.2.
=shell
copy c:\applications\1sen.sdf
copy c:\applications\2sen.sdf
copy c:\applications\3sen.sdf
end
=tsunami-ip
tsunami-ip example
read parameter
e c g lg
values
evalue=0.8
cvalue=0.8
esummary csummary tesum tcsum
end parameter
read response
1sen.sdf use=appl
2sen.sdf use=appl
3sen.sdf use=appl
c:\sensitivity\bnwl2129t3-01sen.sdf use=expt
c:\sensitivity\bnwl2129t3-02sen.sdf use=expt
c:\sensitivity\bnwl2129t3-03sen.sdf use=expt
c:\sensitivity\bnwl2129t3-04sen.sdf use=expt
c:\sensitivity\bnwl2129t3-05sen.sdf use=expt
end response
read reactions
u-235 n,gamma
h-1 elastic
end reactions
end
6.3.1.7. Example PROBLEM
An example problem to demonstrate the use of TSUNAMI-IP is based on the TSUNAMI-1D and TSUNAMI-3D sample problems available with the SCALE distribution. The systems included in the sample problem are not intended to provide a rigorous demonstration of the methodologies of TSUNAMI-IP, but merely to demonstrate how to use the software and read the output. The problem does not correspond to the sample problem distributed with SCALE and is included for illustrative purposes only.
6.3.1.7.1. Input description
The example problem input listing is shown in Example 6.3.3. The input begins with the SCALE shell module, which executes a series of copy commands to move the sensitivity data files required for code execution from the user’s directory (${INPDIR}) to the current working directory. Next TSUNAMI-IP is called with =tsunami-ip. The optional title card is entered as “tsunami-ip example 2.” In the PARAMETER data block, the values table is requested and it will contain \(c_k\), E and G, since c e g are entered on the next line of input. The cutoff value for inclusion of \(c_k\) values in the summary table is set at 0.8 with cvalue=0.8 and the \(c_k\) summary table is requested with csummary. The nuclide-reaction specific integral indices extended \(c_k\) and g are requested with the input c_long and lg. The summary table of g values exceeding the cutoff is requested with lgsum, and the cutoff value for this table is set to 0.75 with lgvalue=0.75. The composite sensitivity profiles for the reactions entered in the REACTIONS input section are requested with prtcomp, and the summary of non-coverage edit is requested with prtnotcv. The uncertainty in the keff values for the experiments and applications due to cross section uncertainties is requested with uncert and the extended uncertainty edits for the applications are requested with uncert_long. The penalty assessment is requested with penalty, the minimum number of experiments exceeding the global integral index; the cutoff value is set to 0 with penminx=0. In this way, the limited number of experiments used in this sample problem will still produce a penalty edit. The detailed edit of the components of the penalty are requested with penlong. The user-defined default value for covariance data are set to 100% uncertainty for cross sections without covariance data with udcov=1.0. The use_dcov input keyword activates the use of the udcov value in the computation of \(c_k\) and the penalty assessment. The use of filenames in place of the titles on the data files is requested with usename.
This input defines two systems as applications in the APPLICATIONS data block. The application systems are defined as the system with sensitivity data in the data files tsunami-3d_k5-1.sdf and tsunami-3d_k5-2.sdf. Three experiments are defined in the EXPERIMENTS data block. These systems have sensitivity data in the data files tsunami-1d1.sdf, tsunami-1d4.sdf, and tsunami-3d_k5-3.sdf.
=shell
cp ${INPDIR}/tsunami-1d1.sdf .
cp ${INPDIR}/tsunami-3d_k5-1.sdf .
cp ${INPDIR}/tsunami-3d_k5-2.sdf .
cp ${INPDIR}/tsunami-1d4.sdf .
cp ${INPDIR}/tsunami-3d_k5-3.sdf .
end
=tsunami-ip
tsunami-ip example 2
read parameter
values
c e g
cvalue=0.8 csummary
c_long
lg lgsum lgvalue=0.75
prtcomp
prtnotcv
uncert uncert_long
penalty penminx=0 penlong
udcov=1.0
use_dcov
use_icov
html usename
end parameter
read applications
tsunami-3d_k5-1.sdf
tsunami-3d_k5-2.sdf
end applications
read experiments
tsunami-1d1.sdf
tsunami-1d4.sdf
tsunami-3d_k5-3.sdf
end experiments
read reactions
u-235 nubar u-238 capture
h-1 total
b-10 capture
end reactions
end
6.3.1.7.2. Output Description
The output listings shown here are for descriptive purposes only. The data generated from an actual execution of this problem may differ from that shown below. The Applications List and the Experiments List provide some details from the sensitivity data files. These include the title on the data files, the filename, the number of energy groups and the number of sensitivity profiles. After the Experiments List, Covariance Warnings are printed during the creation of the working (i.e., problem-dependent) covariance library. The nuclide-reactions pairs present on the sensitivity data files for which covariance data are not available on the coverx file are detailed. This edit is shown in Example 6.3.5.
The next output edit, shown in Example 6.3.6 details the global integral indices for the current application compared to all experiments. The edit is only present if values is entered in the PARAMETER data block. The first row of data in the table displays the application compared to itself. This is useful to confirm the normalization of the integral indices. The data detailing the application compared to itself is not displayed in the HTML output. The next rows display the values of the integral indices requested by the user for each experiment as compared to the current application. The values of the response (i.e., keff or reactivity) for the application and each experiment are also shown in this table. If the sensitivity data were computed with Monte Carlo methods, the statistical uncertainties are shown. In Example 6.3.6, the keff value for tsunami-1d1 does not show a statistical uncertainty because these sensitivity data were computed with TSUNAMI-1D. The integral values of the application compared to tsunami-1d1 do show statistical uncertainties because the application system was computed with TSUNAMI-3D. The column titles “xsec unc %” gives the relative uncertainty in the response due to cross section data uncertainties, in percent.
The next output edit is the extended uncertainty edit for the application and is shown in Example 6.3.7. This edit details the uncertainty in the response due to each nuclide-reaction to nuclide-reaction covariance matrix. Where default or user input values for covariance data are used, they are indicated in the output edit. The total uncertainty can be computed from the individual values by squaring each term, adding the values that had positive signs, subtracting the values that had negative signs, then taking the square root. The edit is present because uncert_long was included in the parameter input.
The next output edit is the extended \(c_k\) edit, and an example edit for Application 1 with Experiment 1 is shown in Example 6.3.8. This edit details the contribution to \(c_k\) due to each nuclide-reaction to nuclide-reaction covariance matrix. Where default or user input values for covariance data are used, they are indicated in the output edit. The edit is present because c_long was included in the parameter input. The \(c_k\) value can be computed by summing the individual values. Similar tables present the output for each application compared to each experiment.
The next output edit includes the nuclide-reaction specific integral indices and is shown in Example 6.3.9. In this sample problem, g indices for fission capture and scatter reactions for each nuclide-reaction pair with a sensitivity coefficient exceeding sencut in magnitude are printed for each experiment compared to the application. The first table of values is for the application compared to itself. This initial edit is useful to test the normalization of the integral indices for the application. This initial table of the application compared with itself is not available in the HTML output. The energy-integrated sensitivity coefficient for the application is also shown for each reaction. Statistical uncertainties in these integral values are not available in this version of TSUNAMI-IP. This table is repeated for each experiment compared to the application. Because some reactions were requested in the REACTIONS section of the input, and the g integral index was computed, a table of the user requested g values for the application compared to each experiment is given. In this sample problem, 10B was not present in the application, even though it was requested in the input. TSUNAMI-IP lists all requested reactions in this table, but leaves the values blank when the nuclide or reaction is not available on the sensitivity data file for the application.
The next output edit, shown in Example 6.3.10, gives the extended penalty assessment for application 1. This edit is present because penlong was entered in the parameter data block. The penalty edit utilizes the user defined default uncertainties defined by udcov because use_dcov was entered in the PARAMETER data block. Nuclide reaction pairs that utilized default uncertainty data are indicated with *. In this case, when the sensitivity coefficients of the application that are not covered by the experiments are propagated to an uncertainty in the application response with the cross section covariance data value of 0.0097% \(\pm\) 0.00007% results. Next a listing of the contributions to the penalty from each nuclide-reaction to nuclide-reaction covariance matrix is given. As with the extended uncertainty edit, the cumulative penalty can be computed from the individual values by squaring each term, adding the values that had positive signs, subtracting the values that had negative signs, then taking the square root.
Each of the previous edits would repeat in the full output listing for each application. The next series of output edits, shown in Example 6.3.11, are the summary edits requested by the user. The \(c_k\) summary table, requested with the keyword csummary, indicates that one experiment has a \(c_k\) value at least as great as 0.8 compared to each application. The g summary table lists the g values that exceed lgvalue for the fission, capture and scatter reactions for each nuclide in the application with an energy-integrated sensitivity coefficient exceeding sencut in magnitude. The value of the sensitivity coefficient for the application is also given. The summary of non-coverage edit is present because prtnotcv was entered in the PARAMETER input data block. This edit lists a number of properties of each reaction input in the REACTIONS input data block. The final summary table is the penalty summary table, which is activated when penalty is listed in the PARAMETER data block. The penalties for all applications identified in the APPLICATIONS input data block are listed in the penalty summary table.
The composite sensitivity data are listed next in the TSUNAMI-IP output as shown in Example 6.3.12 The group-wise values for the composite profiles for each nuclide-reaction pair identified in the REACTIONS data block are listed here for each application identified in the APPLICATIONS data block. If a reaction is requested that is not present in the application, the reaction is left blank in the table, as is the case in this sample problem for 10B capture. The composite sensitivity profiles are also written to a TSUNAMI-B formatted sensitivity data file. In this data file, the composite sensitivity profiles are written for each application in the order they were requested in the REACTIONS data block. If a requested reaction is not available in the application, it is not written to the data file. The application number is listed on the data file in the position of the material number but with a value that is the negative of the application number. All unit and region information are entered as zeros as is the keff value. This data file can be correctly read by Javapeño such that the composite data can be plotted. In Javapeño, the application number appears in the list of available sensitivity profiles identified as “composite.” The application sensitivity data and the composite data as used in the penalty calculation are also included on the composite sensitivity data file. These new data are identified as “application” and “cut composite” respectively.
-----------------------------------------------------------------------------------------------------------------------------------
| |
| TSUNAMI-IP: Tools for Sensitivity and UNcertainty Analysis Methodology Implementation - Indices and Parameters |
| |
| tsunami-ip example 2 |
| |
-----------------------------------------------------------------------------------------------------------------------------------
| |
| TSUNAMI-IP Parameter Table |
| |
| PARAMETER VALUE DESCRIPTION |
| |
| absolute false Print uncertainty values and penalty assessments in absolute format. This is the default |
| format. Relative format can be specified using the "rel" keyword in the APPLICATIONS, |
| EXPERIMENTS, or RESPONSE input blocks. |
| |
| c true Compute c(k) values for each application compared to each experiment. |
| |
| c_long true Produces extended c(k) output edit for each application compared to each experiment. |
| Sets c to true. |
| |
| cechck= .5000 Level of E and c(k) values that trigger the cediff warning. If E or c(k) are below this |
| value, no warning is printed. |
| |
| cediff= .1000 If the E and c(k) values for a given application and experiment differ by more than |
| cediff, a warning message is printed. |
| |
| coverx= 56groupcov7.1 Name of cross section covariance data file. |
| |
| cov_fix false Replace zero and large values on diagonal of cross-section covariance data with user |
| input values and dcov value. |
| |
| cp false Compute and print completeness parameter for each application. |
| |
| csummary true Print summary table of c(k) values that meet or exceed the cvalue limit. Sets c to true. |
| |
| cvalue= .8000 Threshold value of c(k) for inclusion in the summary table. |
| |
| cr false Compute c(r) values for each application compared to each experiment. |
| |
| cr_long false Produces extended c(r) output edit for each application compared to each experiment. |
| Sets cr to true. |
| |
| crsummary false Print summary table of c(r) values that meet or exceed the crvalue limit. Sets cr to |
| true. |
| |
| crvalue= .9000 Threshold value of c(r) for inclusion in the summary table. |
| |
| e true Compute E(sum) values for each application compared to each experiment. |
| |
| esummary false Print summary table of E(sum) values that meet or exceed the evalue limit. Sets e to |
| true. |
| |
| evalue= .9000 Threshold value of E(sum) for inclusion in the summary table. |
| |
| g true Compute G values. |
| |
| gsummary false Print summary table of G values that meet or exceed the gvalue limit. Sets g to true. |
| |
| gvalue= .9000 Threshold value of G for inclusion in summary table. |
| |
| html true Generate output in html format in addition to the standard text output. |
| |
| inptcase false Set the SCALE free form reader to leave the case of the input data as read. |
| |
| large_cov= 10.0000 Cutoff fractional standard deviation value for cov_fix. Covariance data with |
| uncertainties larger than large_cov are replaced with user-defined or default values. |
| |
| lg true Compute g values for fission, capture and scatter for each nuclide for each experiment |
| compared to each application and print them in a table if the application's sensitivity |
| for the corresponding nuclide-reaction pair is greater than or equal to sencut. |
| |
| lgall false Print g values for fission, capture and scatter for all nuclides for all experiments for |
| each application. |
| |
| lggroups false Print a table listing the number of experiments that are at least as sensitive as the |
| application for each group for each reaction requested in the REACTIONS input block. |
| |
| lgsum true Prints a summary table of g for each application for each experiment that exceeds lgvalue |
| for each nuclide's capture, fission and scatter reactions. |
| |
| lgvalue= .7500 Threshold value of g for inclusion in summary table. |
| |
| nixlim= 10 Minimum number of experiments with group-wise values exceeding senfac times the |
| group-wise value for the application for the group-wise value to be added to the |
| completeness parameter. |
| |
| penalty true Creates penalty assessment based on differences in the application's sensitivity profile |
| for a particular nuclide-reaction pair, and the corresponding composite profile for all |
| qualifying experiments. |
| |
| pencut= 0.0000E+00 Cutoff value for excluding sensitivities from the penalty calculation. If the sum of the |
| absolute values of the energy-dependent sensitivity data for a particular nuclide-reaction |
| pair is below this number, the nuclide-reaction pair will not be included in the penalty |
| calculation. |
| |
| penlgv= 0.0000E+00 Use only nuclide-reaction pairs with g values exceeding penlgv in the penalty assessment. |
| |
| penlong true Print detailed edits of components of penalty assessments. |
| |
| penminx= 0 Minimum number of qualifying experiments for each application for penalty calculation. |
| |
| penusec true Use only experiments with c(k) values exceeding cvalue in the penalty assessment. Sets c |
| to true. |
| |
| penusee false Use only experiments with E(sum) values exceeding evalue in the penalty assessment. Sets |
| e to true. |
| |
| penuseg false Use only experiments with G values exceeding gvalue in the penalty assessment. Sets g to |
| true. |
| |
| penwarn false Print list of warning messages noting the experiments and nuclide-reaction pairs that |
| were excluded from the penalty calculation. |
| |
| plot false Produces Ptolemy formatted plot (.ptp) files for integral values and composite |
| sensitivity data. |
| |
| prtcomp true Print "composite" of experiment sensitivity profiles for reactions selected in REACTIONS |
| input block and write data to sensitivity data file. Sets g to true. |
| |
| prtmtrix false Prints directory of data available on the cross section covariance data library. |
| |
| prtnotcv true Print a table summarizing the non-coverage for the nuclide-reaction pairs entered in the |
| REACTIONS input block. Sets g to true. |
| |
| prtparts false Print the components of E and G for fission capture in scatter in the values table. |
| |
| relative true Print uncertainty values and penalty assessments in relative format. This is the default |
| format. Absolute format can be specified using the "abs" keyword in the APPLICATIONS, |
| EXPERIMENTS, or RESPONSE input blocks. |
| |
| return_work_covfalse Option to copy the working covariance data file back to the return directory. |
| |
| sencut= .0100 Cutoff value for ignoring low valued sensitivities in nuclide-reaction specific edit |
| tables. |
| |
| senfac= .9000 Value used in calculation of completeness parameter. Group-wise sensitivity for the |
| application is counted as validated by the experiment if the sensitivity from the |
| experiment is greater than the application sensitivity times senfac. |
| |
| uncert true Computes uncertainty in response due to covariance data. |
| |
| uncert_long true Prints extended table of uncertainty in response due to covariance data. sets uncert to |
| true. |
| |
| use_dcov true Use user-defined default covariance data, udcov, for nuclide reaction pairs not included |
| on the covariance data file. The user-defined data will be used in the fourth penalty |
| assessment as well as the computation of c(k) and uncertainty calculations. |
| |
| udcov= 1.0000 User-defined default value of standard deviation in cross-section data to use for |
| nuclide-reaction pairs for which covariance data are not available on the selected data |
| file. |
| |
| udcov_corr= 1.0000 User-defined default correlation value to use for nuclide-reaction pairs for which |
| covariance data are not available on the selected data file. |
| |
| udcov_corr_typezone User-defined default correlation in cross-section data to use for nuclide-reaction pairs |
| for which covariance data are not available on the selected data file. (long, zone, |
| short) |
| |
| udcov_fast= 0.0000 User-defined default value of standard deviation in cross-section data to use for fast |
| data for nuclide-reaction pairs for which covariance data are not available on the |
| selected data file. |
| |
| udcov_inter= 0.0000 User-defined default value of standard deviation in cross-section data to use for |
| intermediate data for nuclide-reaction pairs for which covariance data are not available |
| on the selected data file. |
| |
| udcov_therm= 0.0000 User-defined default value of standard deviation in cross-section data to use for thermal |
| data for nuclide-reaction pairs for which covariance data are not available on the |
| selected data file. |
| |
| use_diff_groupstrue Allow sensitivity data files to have different energy group structures. |
| |
| use_icov true Use user-defined data input in COVARIANCE input data block in place of udcov value for |
| user input nuclide-reaction pairs that are not on the covariance data file. The |
| user-defined data will be used in the fourth penalty assessment as well as the computation |
| of c(k) and uncertainty calculations. |
| |
| usename true Use the name of the sensitivity data file in place of its title in all output. |
| |
| uslstats false Produces USLSTATS input files for trending analysis with all experiments for each global |
| integral index (c(k), E, and G) requested for each application |
| |
| uslsummary false Produces USLSTATS input files for trending analysis with experiments exceeding cutoff |
| value (cvalue, crvalue, evalue or gvalue) for each global integral index (c(k), E, and G) |
| requested for each application |
| |
| usl_uncert false Includes uncertainty in k-eff due to cross section covariance data from k-eff uncertainty |
| written to USLSTATS input files. |
| |
| usl_p= .9990 Value of P in USLSTATS input files. P is the portion of the population falling above the |
| lower tolerance level |
| |
| usl_1-g= .9500 Value of 1-gamma in USLSTATS input files. 1-gamma is the confidence on the fit. |
| |
| usl_alpha= .9990 Value of alpha in USLSTATS input files. Alpha is the confidence on the proportion of P. |
| |
| usl_xmin= .0000 Value of x(min) in USLSTATS input files. X(min) is the minimum value of the parameter x. |
| |
| usl_xmax= 1.0000 Value of x(max) in USLSTATS input files. X(max) is the maximum value of the parameter x. |
| |
| usl_sigs= -.0030 Value of sigma(s) in USLSTATS input files. If a positive value is entered, it is applied |
| as the total uncertainty for each experiment. If a negative or zero value is entered, it |
| is used as the experimental measurement uncertainty and any Monte Carlo and cross section |
| uncertainties are added quadratically to the absolute value of usl_sigs for each |
| experiment. |
| |
| usl_dkm= .0500 Value of delta-k(m) in USLSTATS input files. Delta-k(m) is the administrative margin |
| used to ensure subcriticality. |
| |
| values true Print all computed "global" indices (E, c(k), and G) in a table for each application. If |
| prtparts is input, also include the partial values. |
| |
-----------------------------------------------------------------------------------------------------------------------------------
| |
| User Requested Reactions for g Analysis |
| |
| NUCLIDE ID NUCLIDE REACTION MT |
| ---------- ------- -------- ---- |
| 92235 u-235 nubar 452 |
| 92238 u-238 capture 101 |
| 1001 h-1 total 1 |
| 5010 b-10 capture 101 |
| |
-----------------------------------------------------------------------------------------------------------------------------------
| |
| HTML Format Options |
| |
| PARAMETER VALUE DESCRIPTION |
| --------- ----- ----------- |
| |
| bg_clr= PapayaWhip Background color |
| |
| h1_clr= Maroon Color used for major headings |
| |
| h2_clr= Navy Color used for sub-headings |
| |
| txt_clr= Black Color for plain text |
| |
| lnk_clr= Navy Color for hyperlinks |
| |
| lnk_dec= none Decoration for hyperlinks (none, underline, overline, line-through, blink) |
| |
| vlnk_clr= Navy Color for visited hyperlinks |
| |
| max_clr= Red Color for maximum values in tables |
| |
| cut_clr= Blue Color for values in tables that exceed cutoff values |
| |
| ud_clr= Blue Color for values in tables that use default covariance data |
| |
| ui_clr= Red Color for values in tables that use user-input covariance data |
| |
| udfix_clr= RoyalBlue Color for values in tables that use default corrected covariance data |
| |
| uifix_clr= Green Color for values in tables that use user-input corrected covariance data |
| |
-----------------------------------------------------------------------------------------------------------------------------------
| |
| Applications List |
| |
| APPLICATION DESCRIPTION |
| |
| 1 Title: tsunami-3d sample 1 |
| Name: tsunami-3d_k5-1, Type: keff , Format: Relative |
| 238 groups, 45 Sensitivity profiles |
| |
| 2 Title: tsunami-3d sample 2 |
| Name: tsunami-3d_k5-2, Type: keff , Format: Relative |
| 238 groups, 406 Sensitivity profiles |
| |
| |
-----------------------------------------------------------------------------------------------------------------------------------
| |
| Experiments List |
| |
| EXPERIMENT DESCRIPTION |
| |
| 1 Title: tsunami-1d sample 1 - keff |
| Name: tsunami-1d1, Type: keff , Format: Relative |
| Filename: tsunami-1d1.sdf |
| 238 groups, 45 Sensitivity profiles |
| |
| 2 Title: tsunami-1d sample 4 |
| Name: tsunami-1d4, Type: keff , Format: Relative |
| Filename: tsunami-1d4.sdf |
| 238 groups, 30 Sensitivity profiles |
| |
| 3 Title: tsunami-3d sample 3 |
| Name: tsunami-3d_k5-3, Type: keff , Format: Relative |
| Filename: tsunami-3d_k5-3.sdf |
| 238 groups, 182 Sensitivity profiles |
| |
| |
------------------------------------------------------------------------------------------
Covariance Warnings in creating working COVERX library
-----------------------------------------------------------------------------------------
Default data will be used for mg n,n'
all= 1.0000 therm= 0.0000 inter = 0.0000 fast= 0.0000 corr= 1.0000 corr_type= zone
Default data will be used for mg n,2n
all= 1.0000 therm= 0.0000 inter = 0.0000 fast= 0.0000 corr= 1.0000 corr_type= zone
Default data will be used for mg n,p
all= 1.0000 therm= 0.0000 inter = 0.0000 fast= 0.0000 corr= 1.0000 corr_type= zone
Default data will be used for mg n,alpha
all= 1.0000 therm= 0.0000 inter = 0.0000 fast= 0.0000 corr= 1.0000 corr_type= zone
Default data will be used for al-27 n,d
all= 1.0000 therm= 0.0000 inter = 0.0000 fast= 0.0000 corr= 1.0000 corr_type= zone
Default data will be used for al-27 n,t
all= 1.0000 therm= 0.0000 inter = 0.0000 fast= 0.0000 corr= 1.0000 corr_type= zone
Default data will be used for si n,2n
all= 1.0000 therm= 0.0000 inter = 0.0000 fast= 0.0000 corr= 1.0000 corr_type= zone
Default data will be used for si n,d
all= 1.0000 therm= 0.0000 inter = 0.0000 fast= 0.0000 corr= 1.0000 corr_type= zone
Default data will be used for s n,n'
all= 1.0000 therm= 0.0000 inter = 0.0000 fast= 0.0000 corr= 1.0000 corr_type= zone
Default data will be used for s n,2n
all= 1.0000 therm= 0.0000 inter = 0.0000 fast= 0.0000 corr= 1.0000 corr_type= zone
Default data will be used for s fission
all= 1.0000 therm= 0.0000 inter = 0.0000 fast= 0.0000 corr= 1.0000 corr_type= zone
Default data will be used for s n,p
all= 1.0000 therm= 0.0000 inter = 0.0000 fast= 0.0000 corr= 1.0000 corr_type= zone
Default data will be used for s n,d
all= 1.0000 therm= 0.0000 inter = 0.0000 fast= 0.0000 corr= 1.0000 corr_type= zone
Default data will be used for s n,t
all= 1.0000 therm= 0.0000 inter = 0.0000 fast= 0.0000 corr= 1.0000 corr_type= zone
Default data will be used for s n,alpha
all= 1.0000 therm= 0.0000 inter = 0.0000 fast= 0.0000 corr= 1.0000 corr_type= zone
Default data will be used for ca n,n'
all= 1.0000 therm= 0.0000 inter = 0.0000 fast= 0.0000 corr= 1.0000 corr_type= zone
Default data will be used for ca n,2n
all= 1.0000 therm= 0.0000 inter = 0.0000 fast= 0.0000 corr= 1.0000 corr_type= zone
Default data will be used for ca n,p
all= 1.0000 therm= 0.0000 inter = 0.0000 fast= 0.0000 corr= 1.0000 corr_type= zone
Default data will be used for ca n,d
all= 1.0000 therm= 0.0000 inter = 0.0000 fast= 0.0000 corr= 1.0000 corr_type= zone
Default data will be used for ca n,t
all= 1.0000 therm= 0.0000 inter = 0.0000 fast= 0.0000 corr= 1.0000 corr_type= zone
-----------------------------------------------------------------
Integral Values for Application #1
-----------------------------------------------------------------
Experiment Type Value s.d. xsec unc % s.d. c(k) s.d. E s.d. G s.d.
------------------------- -------- ---------- --------- ---------- --------- ------ ------ ------ ------ ------ ------
0 tsunami-3d_k5-1 keff 1.0078E+0 1.0000E-3 6.10990E-1 2.1762E-5 1.0000 0.0001 1.0000 0.0005 1.0000 0.0000
1 tsunami-1d1 keff 1.0073E+0 6.06419E-1 0.9999 0.0000 0.9999 0.0003 0.9801 0.0008
2 tsunami-1d4 keff 1.0040E+0 1.42001E+0 0.0988 0.0000 0.0164 0.0000 0.0542 0.0016
3 tsunami-3d_k5-3 keff 9.9575E-1 9.9300E-4 7.48243E-1 1.2500E-4 0.6442 0.0001 0.9647 0.0008 0.6754 0.0026
------------------------------------------------------------------------------------------------------------------------
Extended Uncertainty Edit for Application 1 tsunami-3d1
------------------------------------------------------------------------------------------------------------------------
---------------------------------------------------------------------------------------------------------------------------------
Input covariance file: 56groupcov7.1
Working covariance file: tsunami-ip.wrk
The standard deviation of keff ( % dk/k ) is:
6.110E-01 +/- 2.176E-05 percent
---------------------------------------------------------------------------------------------------------------------------------
* indicates default covariance data
contributions to uncertainty in keff ( % dk/k ) by individual energy covariance matrices:
covariance matrix
nuclide-reaction with nuclide-reaction % dk/k due to this matrix
---------------------- ----------------------- -----------------------------------
u-238 n,gamma u-238 n,gamma 3.8714E-01 +/- 6.2871E-06
u-235 nubar u-235 nubar 2.8509E-01 +/- 7.9001E-06
u-238 n,n' u-238 n,n' 2.2073E-01 +/- 7.7594E-06
u-235 n,gamma u-235 n,gamma 1.6006E-01 +/- 1.7559E-06
f-19 elastic f-19 elastic 1.3624E-01 +/- 5.0707E-06
u-238 elastic u-238 n,n' -1.2828E-01 +/- 1.7674E-06
u-235 fission u-235 n,gamma 1.2387E-01 +/- 8.3076E-07
u-235 fission u-235 fission 1.2134E-01 +/- 1.2085E-06
h-1 elastic h-1 elastic 1.1972E-01 +/- 2.1606E-06
f-19 elastic f-19 n,n' -1.1793E-01 +/- 3.1965E-06
f-19 n,n' f-19 n,n' 1.1286E-01 +/- 3.8652E-06
u-235 chi u-235 chi 8.8178E-02 +/- 1.5583E-05
u-238 elastic u-238 elastic 6.9520E-02 +/- 1.1586E-06
u-238 nubar u-238 nubar 5.8614E-02 +/- 5.4192E-07
h-1 n,gamma h-1 n,gamma 5.0829E-02 +/- 1.6728E-07
u-238 elastic u-238 n,gamma 5.0286E-02 +/- 1.7408E-06
f-19 n,alpha f-19 n,alpha 1.9795E-02 +/- 1.0127E-07
u-238 fission u-238 fission 1.7394E-02 +/- 3.4394E-08
c elastic c elastic 1.5520E-02 +/- 5.5754E-08
u-238 n,2n u-238 n,2n 1.3981E-02 +/- 1.2056E-07
f-19 n,gamma f-19 n,gamma 9.7994E-03 +/- 6.0845E-09
c n,n' c elastic -9.0325E-03 +/- 3.2330E-08
c n,n' c n,n' 8.6479E-03 +/- 5.6289E-08
f-19 elastic f-19 n,alpha 6.6750E-03 +/- 1.2243E-08
------------------------------------------------------------------------------------------------------------------------
Extended c(k) Edit for Application 1 tsunami-3d1 with Experiment 1 tsunami-1d1
------------------------------------------------------------------------------------------------------------------------
---------------------------------------------------------------------------------------------------------------------------------
the c(k) value is:
1.0000 +/- 0.0001
---------------------------------------------------------------------------------------------------------------------------------
Input covariance file: 56groupcov7.1
Working covariance file: tsunami-ip.wrk
* indicates default covariance data
contributions to c(k) by individual energy covariance matrices
the c(k) value is the sum of the individual contributions
covariance matrix
nuclide-reaction with nuclide-reaction c(k) contribution from this matrix individual c(k)
---------------------- ----------------------- ----------------------------------- -----------------------------
u-238 n,gamma u-238 n,gamma 4.0149E-01 +/- 1.4552E-05 1.0000E+00 +/- 3.6246E-05
u-235 nubar u-235 nubar 2.1772E-01 +/- 1.8286E-05 1.0000E+00 +/- 8.3989E-05
u-238 n,n' u-238 n,n' 1.3051E-01 +/- 1.7960E-05 1.0000E+00 +/- 1.3761E-04
u-235 n,gamma u-235 n,gamma 6.8631E-02 +/- 4.0642E-06 1.0000E+00 +/- 5.9217E-05
f-19 elastic f-19 elastic 4.9723E-02 +/- 1.1737E-05 1.0000E+00 +/- 2.3604E-04
u-235 fission u-235 fission 3.9440E-02 +/- 2.7972E-06 1.0000E+00 +/- 7.0924E-05
h-1 elastic h-1 elastic 3.8391E-02 +/- 5.0010E-06 1.0000E+00 +/- 1.3026E-04
f-19 n,n' f-19 n,n' 3.4122E-02 +/- 8.9464E-06 1.0000E+00 +/- 2.6219E-04
u-238 elastic u-238 n,n' -2.2039E-02 +/- 2.8927E-06
u-238 n,n' u-238 elastic -2.2039E-02 +/- 2.8927E-06
u-235 chi u-235 chi 2.0828E-02 +/- 3.6069E-05 1.0000E+00 +/- 1.7317E-03
u-235 fission u-235 n,gamma 2.0550E-02 +/- 1.3597E-06
u-235 n,gamma u-235 fission 2.0550E-02 +/- 1.3597E-06
f-19 elastic f-19 n,n' -1.8628E-02 +/- 5.2316E-06
f-19 n,n' f-19 elastic -1.8628E-02 +/- 5.2316E-06
u-238 elastic u-238 elastic 1.2947E-02 +/- 2.6818E-06 1.0000E+00 +/- 2.0714E-04
u-238 nubar u-238 nubar 9.2031E-03 +/- 1.2543E-06 1.0000E+00 +/- 1.3629E-04
h-1 n,gamma h-1 n,gamma 6.9207E-03 +/- 3.8719E-07 1.0000E+00 +/- 5.5946E-05
u-238 elastic u-238 n,gamma 3.3869E-03 +/- 2.8491E-06
u-238 n,gamma u-238 elastic 3.3869E-03 +/- 2.8491E-06
f-19 n,alpha f-19 n,alpha 1.0496E-03 +/- 2.3439E-07 1.0000E+00 +/- 2.2331E-04
u-238 fission u-238 fission 8.1042E-04 +/- 7.9610E-08 1.0000E+00 +/- 9.8233E-05
c elastic c elastic 6.4522E-04 +/- 1.2905E-07 1.0000E+00 +/- 2.0001E-04
u-238 n,2n u-238 n,2n 5.2362E-04 +/- 2.7904E-07 1.0000E+00 +/- 5.3290E-04
----------------------------------------------
NUCLIDE-REACTION SPECIFIC INTEGRAL INDICES
----------------------------------------------
nuclide reaction sensitivity g
------------- -------- ----------- ------
h-1 capture -1.02E-01 1.0000
h-1 scatter 3.22E-01 1.0000
c scatter 2.48E-02 1.0000
f-19 scatter 4.70E-02 1.0000
u-235 fission 3.63E-01 1.0000
u-235 capture -1.13E-01 1.0000
u-238 fission 3.35E-02 1.0000
u-238 capture -2.87E-01 1.0000
u-238 scatter 4.88E-02 1.0000
nuclide reaction sensitivity g
------------- -------- ----------- ------
h-1 capture -1.02E-01 0.9971
h-1 scatter 3.22E-01 0.9413
c scatter 2.48E-02 0.9491
f-19 scatter 4.70E-02 0.9511
u-235 fission 3.63E-01 0.9998
u-235 capture -1.13E-01 0.9967
u-238 fission 3.35E-02 0.9978
u-238 capture -2.87E-01 0.9960
u-238 scatter 4.88E-02 0.9773
nuclide reaction sensitivity g
------------- -------- ----------- ------
h-1 capture -1.02E-01 0.0000
h-1 scatter 3.22E-01 0.0000
c scatter 2.48E-02 0.0000
f-19 scatter 4.70E-02 0.0000
u-235 fission 3.63E-01 0.0143
u-235 capture -1.13E-01 0.0072
u-238 fission 3.35E-02 0.9996
u-238 capture -2.87E-01 0.0481
u-238 scatter 4.88E-02 0.4112
----------------------------------------------------------------------------------------------------------------------------
Values for reactions with sensitivities greater than 1.00E-02 for application tsunami-3d_k5-1 with experiment tsunami-
----------------------------------------------------------------------------------------------------------------------------
nuclide reaction sensitivity g
------------- -------- ----------- ------
h-1 capture -1.02E-01 0.9994
h-1 scatter 3.22E-01 0.7755
c scatter 2.48E-02 0.0076
f-19 scatter 4.70E-02 0.0000
u-235 fission 3.63E-01 0.8622
u-235 capture -1.13E-01 0.9364
u-238 fission 3.35E-02 0.7276
u-238 capture -2.87E-01 0.3955
u-238 scatter 4.88E-02 0.4822
------------------------------------------------
User requested g values for tsunami-3d_k5-1
------------------------------------------------
u-235 u-238 h-1 b-10
Experiment nubar capture total capture
---------------- -------- -------- -------- --------
tsunami-1d1 0.9991 0.9960 0.9445
tsunami-1d4 0.0095 0.0481 0.0000
tsunami-3d_k5-3 0.9403 0.3955 0.8433
------------------------------------------------------------------------------------------------------------------------
Penalty Assessment for Application 1 tsunami-3d1
------------------------------------------------------------------------------------------------------------------------
---------------------------------------------------------------------------------------------------------------------------------
the standard deviation of keff due to uncovered sensitivity data is:
9.747E-03 +/- 6.584E-05 ( % dk/k )
---------------------------------------------------------------------------------------------------------------------------------
Input covariance file: 56groupcov7.1
Working covariance file: tsunami-ip.wrk
* indicates default covariance data
contributions to uncertainty in keff ( % dk/k ) by individual energy covariance matrices:
covariance matrix
nuclide-reaction with nuclide-reaction % dk/k due to this matrix
---------------------- ----------------------- -----------------------------------
u-238 n,n' u-238 n,n' 7.5820E-03 +/- 1.8458E-05
h-1 elastic h-1 elastic 4.1794E-03 +/- 4.5264E-06
f-19 elastic f-19 elastic 3.9736E-03 +/- 1.0132E-05
u-238 elastic u-238 n,n' -3.6600E-03 +/- 2.8032E-05
u-235 chi u-235 chi 3.6588E-03 +/- 4.5806E-05
f-19 n,n' f-19 n,n' 2.9919E-03 +/- 5.7578E-06
f-19 elastic f-19 n,n' -2.9052E-03 +/- 3.0295E-05
u-238 n,gamma u-238 n,gamma 1.2407E-03 +/- 1.3961E-06
u-238 elastic u-238 elastic 1.1105E-03 +/- 9.2475E-07
u-238 elastic u-238 n,gamma 5.6491E-04 +/- 5.8032E-06
c elastic c elastic 4.8848E-04 +/- 1.0550E-07
u-235 n,gamma u-235 n,gamma 4.4181E-04 +/- 2.6790E-07
u-238 n,2n u-238 n,2n 3.2664E-04 +/- 1.8609E-07
u-238 chi u-238 chi 2.5281E-04 +/- 1.9474E-07
u-235 nubar u-235 nubar 1.7677E-04 +/- 2.0085E-07
c n,n' c elastic -1.7595E-04 +/- 2.8179E-07
c n,n' c n,n' 1.7558E-04 +/- 7.7046E-08
h-1 n,gamma h-1 n,gamma 1.4297E-04 +/- 2.6298E-08
u-238 nubar u-238 nubar 9.8421E-05 +/- 3.5167E-08
f-19 n,alpha f-19 n,alpha 6.4564E-05 +/- 1.1052E-08
u-238 elastic u-238 n,2n -4.7329E-05 +/- 2.0045E-08
u-235 n,n' u-235 n,n' 4.5469E-05 +/- 5.5758E-10
f-19 elastic f-19 n,alpha 4.2446E-05 +/- 2.0604E-08
------------------------------
c(k) SUMMARY TABLE
------------------------------
application # 1 tsunami-3d_k5-1 has 1 c(k) values >= 0.8000
1 tsunami-1d1 0.9999 +/- 0.0000
application # 2 tsunami-3d_k5-2 has 1 c(k) values >= 0.8000
1 tsunami-1d1 0.8175 +/- 0.0001
-----------------------------------------------------------------
Summary of Integral Values Meeting Acceptance Criteria
-----------------------------------------------------------------
Application c(k) >= 0.8000
----------------- ----------------
tsunami-3d_k5-1 1
tsunami-3d_k5-2 1
----------------
g SUMMARY TABLE
----------------
application #1 tsunami-3d_k5-1
h-1 capture ( sensitivity = -1.0173E-01 +/- 1.2526E-05 ) has 2 g values >= 0.7500
1 tsunami-1d1 0.9971
3 tsunami-3d_k5-3 0.9994
h-1 scatter ( sensitivity = 3.2202E-01 +/- 1.9772E-04 ) has 2 g values >= 0.7500
1 tsunami-1d1 0.9413
3 tsunami-3d_k5-3 0.7755
c scatter ( sensitivity = 2.4841E-02 +/- 1.4343E-05 ) has 1 g values >= 0.7500
1 tsunami-1d1 0.9491
f-19 scatter ( sensitivity = 4.6979E-02 +/- 2.0034E-05 ) has 1 g values >= 0.7500
1 tsunami-1d1 0.9511
u-235 fission ( sensitivity = 3.6286E-01 +/- 5.2950E-05 ) has 2 g values >= 0.7500
1 tsunami-1d1 0.9998
3 tsunami-3d_k5-3 0.8622
u-235 capture ( sensitivity = -1.1286E-01 +/- 1.2704E-05 ) has 2 g values >= 0.7500
1 tsunami-1d1 0.9967
3 tsunami-3d_k5-3 0.9364
u-238 fission ( sensitivity = 3.3498E-02 +/- 5.4895E-06 ) has 2 g values >= 0.7500
1 tsunami-1d1 0.9978
2 tsunami-1d4 0.9996
u-238 capture ( sensitivity = -2.8728E-01 +/- 2.1554E-05 ) has 1 g values >= 0.7500
1 tsunami-1d1 0.9960
application #2 tsunami-3d_k5-2
h-1 capture ( sensitivity = -1.9682E-01 +/- 5.1773E-05 ) has 1 g values >= 0.7500
3 tsunami-3d_k5-3 0.9921
h-1 scatter ( sensitivity = 3.5602E-01 +/- 1.9469E-03 ) has 1 g values >= 0.7500
3 tsunami-3d_k5-3 0.9160
o-16 scatter ( sensitivity = 6.9366E-02 +/- 9.7179E-05 ) has 1 g values >= 0.7500
3 tsunami-3d_k5-3 0.9229
u-235 fission ( sensitivity = 3.2845E-01 +/- 1.3867E-04 ) has 2 g values >= 0.7500
1 tsunami-1d1 0.9687
3 tsunami-3d_k5-3 0.9751
u-235 capture ( sensitivity = -1.1817E-01 +/- 3.5477E-05 ) has 2 g values >= 0.7500
1 tsunami-1d1 0.9000
3 tsunami-3d_k5-3 0.9841
u-238 fission ( sensitivity = 2.4663E-02 +/- 9.8534E-06 ) has 3 g values >= 0.7500
1 tsunami-1d1 1.0000
2 tsunami-1d4 0.9998
3 tsunami-3d_k5-3 0.9813
u-238 capture ( sensitivity = -1.1333E-01 +/- 2.3953E-05 ) has 2 g values >= 0.7500
1 tsunami-1d1 1.0000
3 tsunami-3d_k5-3 0.9868
------------------------------
SUMMARY OF NON-COVERAGE
------------------------------
------------------------------------------------------------------------------------------------------------------------
Non-coverage for user-requested reactions for tsunami-3d_k5-1
------------------------------------------------------------------------------------------------------------------------
u-235 u-238 h-1 b-10
nubar capture total capture
---------------------------------- ---------------- ---------------- ---------------- ----------------
Number of groups not covered 63 139 96
Sum of sensitivities not covered 3.9598E-04 -1.1226E-03 8.0684E-03
Group with largest sensitivity 225 225 225
Largest sensitivity group value 2.0287E-01 -2.8184E-02 -2.1178E-02
Best experiment for max group tsunami-1d1 tsunami-1d1 tsunami-3d_k5-3
Group sensitivity for best exp 2.0296E-01 -2.8154E-02 -3.8871E-02
------------------------------------------------------------------------------------------------------------------------
Non-coverage for user-requested reactions for tsunami-3d_k5-2
------------------------------------------------------------------------------------------------------------------------
u-235 u-238 h-1 b-10
nubar capture total capture
---------------------------------- ---------------- ---------------- ---------------- ----------------
Number of groups not covered 22 0 91
Sum of sensitivities not covered 3.7824E-03 0.0000E+00 -1.2898E-03
Group with largest sensitivity 225 225 225
Largest sensitivity group value 1.9220E-01 -1.3369E-02 -4.6670E-02
Best experiment for max group tsunami-1d1 tsunami-1d1 tsunami-3d_k5-3
Group sensitivity for best exp 2.0296E-01 -2.8154E-02 -3.8871E-02
------------------------------
PENALTY SUMMARY TABLE
------------------------------
Standard Deviation in Application Response Due to Uncovered Sensitivity Coefficients
Application Type Penalty SD Units
---------------- -------- ------------------------ --------
tsunami-3d_k5-1 keff 9.7475E-3 +/- 6.5845E-5 % dk/k
tsunami-3d_k5-2 keff 2.3500E-1 +/- 1.2902E-4 % dk/k
------------------------------
COMPOSITE SENSITIVITY DATA
------------------------------
------------------------------------------------------------------
Composite of user-requested reactions for tsunami-3d_k5-1
------------------------------------------------------------------
u-235 u-238 h-1 b-10
Group nubar capture total capture
-------- ---------------- ---------------- ---------------- ----------------
1 0.0000E+00 0.0000E+00 0.0000E+00
2 8.3121E-08 -2.1269E-10 -4.8103E-07
3 2.8391E-07 -1.1742E-09 -1.2188E-06
4 5.1635E-07 -2.9305E-09 -1.4983E-06
5 7.6669E-07 -6.5763E-09 -3.0912E-06
6 1.2087E-05 -1.3860E-07 -4.2146E-05
7 3.2436E-05 -2.5827E-07 -7.0385E-05
8 9.4203E-05 -1.5178E-06 2.2338E-04
9 1.7111E-04 -9.8460E-06 2.6160E-03
10 9.5881E-05 -9.6314E-06 1.4498E-03
11 4.6498E-04 -8.6324E-05 6.0348E-03
12 3.3668E-04 -1.0881E-04 4.1605E-03
13 1.0308E-04 -4.2404E-05 1.5022E-03
14 4.9039E-04 -2.8588E-04 6.9219E-03
15 4.1834E-04 -3.6981E-04 6.6199E-03
16 1.3739E-04 -1.5075E-04 2.9036E-03
17 6.1269E-05 -7.3358E-05 1.2879E-03
18 5.3656E-05 -6.8243E-05 1.1029E-03
19 9.3382E-05 -1.2782E-04 1.8069E-03
20 7.2274E-05 -1.0611E-04 1.7937E-03
21 1.6121E-04 -2.6852E-04 3.6259E-03
22 1.5722E-04 -2.9405E-04 3.6598E-03
23 1.6032E-04 -3.2036E-04 3.6221E-03
24 3.6667E-05 -7.5670E-05 8.7492E-04
25 4.6036E-05 -9.5868E-05 1.1263E-03
26 2.4917E-05 -5.2550E-05 5.7950E-04
27 7.3525E-05 -1.5597E-04 1.6697E-03
28 1.3207E-04 -2.7919E-04 3.1426E-03
29 1.4917E-04 -3.1499E-04 3.3212E-03
30 1.9524E-05 -4.0694E-05 4.7128E-04
31 1.5260E-04 -3.1113E-04 3.5947E-03
32 5.9914E-05 -1.2004E-04 1.0086E-03
33 5.4703E-05 -1.0849E-04 1.0609E-03
34 1.2067E-04 -2.3813E-04 2.0801E-03
35 7.4939E-05 -1.4661E-04 1.3644E-03
36 7.5277E-05 -1.4645E-04 1.3422E-03
37 4.5884E-05 -8.8788E-05 6.8414E-04
38 5.1306E-05 -9.8727E-05 9.3662E-04
39 1.9597E-04 -3.7831E-04 3.5664E-03
40 1.8431E-04 -3.6387E-04 1.8026E-03
41 2.8555E-04 -5.5723E-04 4.2242E-03
42 2.6119E-04 -5.3510E-04 4.7957E-03
43 1.3711E-04 -2.9893E-04 1.8811E-03
44 1.9055E-04 -4.3852E-04 2.1220E-03
45 1.2409E-04 -3.1979E-04 1.1164E-03
46 3.6535E-05 -9.7941E-05 4.6072E-04
47 8.7249E-05 -2.3882E-04 8.8505E-04
48 2.5653E-05 -6.9180E-05 3.5130E-04
49 1.6739E-04 -5.0475E-04 2.2125E-03
50 1.1716E-04 -3.9760E-04 1.5983E-03
51 2.4526E-05 -9.6985E-05 1.5119E-04
52 6.4964E-05 -2.6803E-04 7.1621E-04
53 3.3839E-04 -1.4814E-03 4.7531E-03
54 1.3114E-04 -6.1020E-04 1.8245E-03
55 3.5173E-04 -1.7047E-03 5.1134E-03
56 2.4929E-04 -1.2514E-03 3.3044E-03
57 3.1251E-04 -1.5616E-03 3.2685E-03
58 1.7934E-04 -7.8059E-04 1.6846E-03
59 3.3768E-04 -1.5847E-03 3.0949E-03
60 5.7218E-04 -2.6453E-03 4.6208E-03
61 6.2408E-05 -2.1987E-04 2.8987E-04
62 3.4221E-04 -1.6321E-03 2.6850E-03
63 2.5176E-04 -1.0549E-03 1.7729E-03
64 2.1465E-04 -9.5589E-04 1.3821E-03
65 7.3062E-05 -3.8956E-04 5.2412E-04
66 3.7161E-04 -1.4107E-03 2.3877E-03
67 3.2093E-04 -1.3730E-03 1.6616E-03
68 5.8195E-05 -2.1647E-04 3.7317E-04
69 6.6370E-04 -2.5516E-03 3.5190E-03
70 4.5913E-04 -2.0723E-03 2.4579E-03
71 9.7426E-04 -4.1869E-03 4.9772E-03
72 1.0966E-04 -5.8274E-05 1.8589E-04
73 7.1828E-04 -2.6476E-03 2.7657E-03
74 2.4703E-03 -6.6484E-03 7.4726E-03
75 2.6381E-04 -1.3566E-03 1.1842E-03
76 1.1644E-03 -1.9283E-03 2.1884E-03
77 9.0059E-04 -1.9983E-03 2.0712E-03
78 1.3932E-05 -1.7300E-03 6.6140E-04
79 4.7174E-04 -3.7028E-04 5.4439E-04
80 1.2166E-04 -2.3242E-03 1.2190E-03
81 2.6867E-03 -3.1503E-03 5.5591E-03
82 1.7934E-04 -7.5046E-05 4.2063E-04
83 2.9797E-04 -3.1650E-03 1.6682E-03
84 1.8506E-04 -1.7962E-04 6.9532E-04
85 3.8625E-04 -5.4319E-03 3.0824E-03
86 6.6282E-04 -2.9944E-04 1.2789E-03
87 8.2978E-04 -4.5155E-04 1.1380E-03
88 1.1337E-04 -2.5098E-03 1.9443E-03
89 2.1511E-04 -6.1408E-05 6.1627E-04
90 5.9065E-04 -6.5874E-05 5.6950E-04
91 5.4779E-04 -3.0607E-04 1.0246E-03
92 3.7293E-05 -5.7959E-03 3.1081E-03
93 2.7815E-04 -6.1499E-04 8.7462E-04
94 2.5372E-04 -4.8609E-05 1.2161E-04
95 1.8329E-03 -8.5639E-05 4.8164E-04
96 3.9099E-04 -1.8614E-05 1.2128E-04
97 7.1230E-04 -1.9738E-05 1.5539E-04
98 2.4608E-04 -3.5551E-05 3.8414E-04
99 2.2019E-04 -1.5836E-05 1.7509E-04
100 3.1350E-04 -2.7103E-05 3.2313E-04
101 2.9855E-04 -6.8308E-05 5.6510E-04
102 3.5827E-04 -5.9559E-05 4.5259E-04
103 2.1822E-04 -1.0055E-04 8.6367E-04
104 3.8254E-04 -1.3022E-04 7.8610E-04
105 2.6271E-04 -2.6041E-04 7.6712E-04
106 4.5665E-04 -1.6304E-04 3.4290E-04
107 1.3250E-04 -7.8605E-04 1.3860E-03
108 5.8291E-05 -3.1200E-03 3.3141E-03
109 2.5917E-04 -9.6788E-03 5.0386E-03
110 1.5727E-03 -9.5678E-04 5.9505E-04
111 5.1745E-04 -3.9567E-04 5.1849E-04
112 3.3689E-04 -1.4825E-04 2.9832E-04
113 5.5476E-04 -2.9394E-04 5.5464E-04
114 2.2143E-05 -6.9191E-05 1.6138E-04
115 1.9064E-04 -1.4094E-04 5.3270E-04
116 5.0989E-04 -2.2832E-04 1.2496E-03
117 1.1009E-03 -3.0055E-04 1.6754E-03
118 1.4392E-03 -8.5266E-04 3.3368E-03
119 2.4313E-04 -7.0828E-03 6.5606E-03
120 1.7047E-04 -8.4025E-03 5.1957E-03
121 2.2684E-03 -1.6808E-03 1.4346E-03
122 2.0681E-04 -3.2469E-04 4.1713E-04
123 5.3283E-04 -5.4572E-04 9.2483E-04
124 5.6983E-04 -2.1805E-04 5.6677E-04
125 3.4396E-04 -1.5890E-04 4.4437E-04
126 2.2707E-04 -1.1013E-04 4.5908E-04
127 8.3158E-04 -9.6956E-05 1.8533E-04
128 4.5995E-04 -1.2467E-04 4.7548E-04
129 1.7971E-03 -1.4955E-04 8.4077E-04
130 3.2110E-04 -6.5461E-05 9.1569E-04
131 6.1328E-04 -8.3373E-04 1.7860E-03
132 1.2347E-03 -3.2726E-04 1.4198E-03
133 4.8042E-03 -7.7205E-04 2.9930E-03
134 1.9295E-04 -3.7846E-03 8.5681E-03
135 3.2715E-04 -2.2262E-03 2.7745E-03
136 6.6065E-05 -7.5091E-03 6.8661E-03
137 1.3387E-05 -3.9274E-03 3.1875E-03
138 5.5488E-04 -6.5404E-03 5.8490E-03
139 3.2411E-04 -2.8026E-03 2.5508E-03
140 3.8712E-04 -2.8312E-03 2.6262E-03
141 1.4925E-04 -9.6337E-04 1.3378E-03
142 1.4422E-04 -4.5295E-04 1.1235E-03
143 1.5490E-04 -1.2351E-03 2.1938E-03
144 6.4744E-05 -3.3445E-04 7.5336E-04
145 8.2229E-04 -2.7165E-04 6.6269E-04
146 6.7978E-04 -4.1042E-04 7.9458E-04
147 3.2938E-04 -1.1761E-04 1.9318E-04
148 1.0859E-04 -5.9175E-05 1.3243E-04
149 5.9627E-05 -3.7701E-05 9.7112E-05
150 1.7015E-04 -1.2409E-04 2.8871E-04
151 1.3042E-04 -1.2721E-04 3.4573E-04
152 7.9303E-05 -1.3007E-04 3.5622E-04
153 8.0252E-05 -1.3289E-04 3.9765E-04
154 9.3218E-05 -1.3512E-04 3.3495E-04
155 9.3312E-05 -1.2361E-04 2.5897E-04
156 9.2711E-05 -1.1448E-04 3.3308E-04
157 1.1417E-04 -1.3297E-04 3.5854E-04
158 1.2864E-04 -1.3720E-04 3.7838E-04
159 3.0061E-04 -1.9035E-04 6.5148E-04
160 1.2085E-04 -9.9337E-05 3.2785E-04
161 1.4351E-04 -1.3909E-04 3.1532E-04
162 1.6682E-04 -1.6273E-04 4.2556E-04
163 1.8495E-04 -1.7393E-04 3.0967E-04
164 2.0345E-04 -1.8509E-04 4.6764E-04
165 2.2132E-04 -1.9666E-04 4.2683E-04
166 1.3542E-04 -1.1483E-04 2.8152E-04
167 1.5182E-04 -1.2110E-04 3.3331E-04
168 1.7660E-04 -1.2685E-04 2.7533E-04
169 2.1245E-04 -1.3208E-04 3.0394E-04
170 2.6320E-04 -1.3841E-04 2.7320E-04
171 1.9419E-04 -7.1841E-05 1.6409E-04
172 2.7519E-04 -7.3711E-05 1.4999E-04
173 3.8550E-04 -7.5674E-05 1.4648E-04
174 5.0044E-04 -7.7661E-05 8.4438E-05
175 2.2717E-04 -3.1699E-05 3.8770E-05
176 2.2913E-04 -3.1410E-05 2.9601E-05
177 2.3709E-04 -3.2293E-05 4.2854E-05
178 2.4257E-04 -3.2709E-05 3.4193E-05
179 2.3003E-04 -3.2775E-05 2.9616E-05
180 2.2349E-04 -3.2936E-05 3.7056E-05
181 2.2113E-04 -3.3917E-05 3.6280E-05
182 2.1354E-04 -3.4383E-05 3.6401E-05
183 2.0719E-04 -3.4859E-05 5.0084E-05
184 2.0168E-04 -3.5283E-05 4.0429E-05
185 1.9414E-04 -3.5733E-05 5.6482E-05
186 1.8530E-04 -3.6287E-05 5.3350E-05
187 1.8060E-04 -3.6876E-05 4.6883E-05
188 1.7542E-04 -3.7408E-05 4.8868E-05
189 1.7875E-04 -3.8136E-05 1.7370E-05
190 4.1786E-04 -9.7698E-05 1.3305E-04
191 4.0193E-04 -1.0043E-04 1.4039E-04
192 4.0255E-04 -1.0398E-04 1.5921E-04
193 3.9780E-04 -1.0814E-04 1.2778E-04
194 8.0388E-04 -2.3098E-04 2.8322E-04
195 8.5863E-04 -2.5171E-04 1.3920E-04
196 9.2365E-04 -2.7401E-04 3.7939E-04
197 1.0456E-03 -3.0662E-04 4.0857E-04
198 1.1698E-03 -3.3550E-04 4.2186E-04
199 6.4963E-04 -1.8310E-04 2.6318E-04
200 7.1981E-04 -1.9880E-04 2.2883E-04
201 1.6158E-03 -4.2613E-04 3.6152E-04
202 2.0254E-03 -4.9966E-04 2.8211E-04
203 2.4707E-03 -5.6340E-04 2.8843E-04
204 3.3624E-03 -6.7630E-04 2.8969E-04
205 2.1844E-03 -3.8558E-04 -8.4664E-05
206 2.7103E-03 -4.3373E-04 1.1227E-04
207 3.2507E-03 -4.6733E-04 0.0000E+00
208 4.1765E-03 -5.5377E-04 1.9915E-05
209 4.6469E-03 -6.0428E-04 -6.5711E-05
210 5.4292E-03 -7.4815E-04 -5.8270E-05
211 5.9319E-03 -8.9850E-04 -4.1482E-05
212 6.8645E-03 -1.1352E-03 3.6827E-06
213 9.0697E-03 -1.5740E-03 9.5084E-05
214 1.3105E-02 -2.3116E-03 -6.2132E-04
215 2.0944E-02 -3.6390E-03 0.0000E+00
216 3.7206E-02 -6.2563E-03 -2.7238E-03
217 2.2374E-02 -3.6629E-03 -1.8224E-03
218 2.9838E-02 -4.7756E-03 -3.1442E-03
219 3.8551E-02 -6.0551E-03 -2.1233E-03
220 4.9816E-02 -7.6542E-03 -4.8489E-03
221 6.4973E-02 -9.7526E-03 -4.3945E-03
222 8.2484E-02 -1.2128E-02 -8.4682E-03
223 1.0354E-01 -1.4855E-02 -9.5965E-03
224 5.5503E-02 -7.8609E-03 -5.7398E-03
225 2.0287E-01 -2.8154E-02 -2.1178E-02
226 3.2324E-02 -4.4137E-03 -3.9011E-03
227 2.9803E-02 -4.0284E-03 -3.8972E-03
228 1.0735E-02 -1.4428E-03 -1.2831E-03
229 9.7841E-03 -1.3106E-03 -1.2401E-03
230 4.4405E-03 -5.9294E-04 -4.6629E-04
231 4.0564E-03 -5.4338E-04 -4.6124E-04
232 3.6568E-03 -4.8680E-04 -4.1461E-04
233 1.9509E-03 -2.5916E-04 -2.2200E-04
234 1.1574E-03 -1.5422E-04 -1.3472E-04
235 1.3127E-03 -1.7411E-04 -1.4945E-04
236 1.1070E-03 -1.4705E-04 -1.1858E-04
237 1.2172E-03 -1.6145E-04 -1.3854E-04
238 1.0399E-04 -1.3798E-05 -1.1822E-05
------------------------------------------------------------------
Composite of user-requested reactions for tsunami-3d_k5-2
------------------------------------------------------------------
u-235 u-238 h-1 b-10
Group nubar capture total capture
-------- ---------------- ---------------- ---------------- ----------------
1 0.0000E+00 0.0000E+00 0.0000E+00
2 1.2519E-07 -8.6672E-11 0.0000E+00
3 3.3421E-07 -4.3407E-10 4.2389E-07
4 7.2762E-07 -1.3902E-09 6.8757E-07
5 1.3021E-06 -3.8278E-09 2.5489E-06
6 1.4922E-05 -5.6904E-08 4.6252E-05
7 4.7469E-05 -1.3730E-07 2.0214E-04
8 1.4446E-04 -8.9470E-07 1.0914E-03
9 2.8286E-04 -6.6636E-06 4.8035E-03
10 1.5596E-04 -6.5378E-06 2.9555E-03
11 7.5246E-04 -5.9712E-05 1.3113E-02
12 5.8627E-04 -7.9447E-05 1.2557E-02
13 1.7542E-04 -3.0813E-05 4.0300E-03
14 7.3736E-04 -1.8899E-04 1.5889E-02
15 6.2601E-04 -2.4990E-04 1.3073E-02
16 2.0457E-04 -1.0331E-04 4.7828E-03
17 9.0359E-05 -4.9631E-05 2.1468E-03
18 7.3377E-05 -4.3188E-05 1.6032E-03
19 1.3573E-04 -8.6428E-05 2.6278E-03
20 1.1161E-04 -7.6638E-05 2.4900E-03
21 2.2033E-04 -1.7115E-04 4.9231E-03
22 1.8239E-04 -1.6205E-04 3.4382E-03
23 1.9578E-04 -1.8691E-04 3.6221E-03
24 4.7147E-05 -4.5670E-05 1.1450E-03
25 6.3898E-05 -6.1821E-05 1.6513E-03
26 3.7065E-05 -3.6333E-05 9.5190E-04
27 1.1856E-04 -1.1649E-04 3.1639E-03
28 2.2510E-04 -2.2146E-04 5.6953E-03
29 2.1545E-04 -2.1077E-04 6.2475E-03
30 2.7193E-05 -2.6428E-05 7.8781E-04
31 2.1388E-04 -2.0401E-04 6.4215E-03
32 8.3295E-05 -7.7147E-05 2.3947E-03
33 7.1583E-05 -6.6441E-05 2.0713E-03
34 1.5905E-04 -1.4732E-04 4.5168E-03
35 9.2541E-05 -8.5375E-05 2.5578E-03
36 8.1119E-05 -7.5870E-05 1.5017E-03
37 4.9873E-05 -4.6393E-05 6.8414E-04
38 6.3817E-05 -5.9216E-05 9.7337E-04
39 2.7366E-04 -2.5218E-04 6.3032E-03
40 2.7217E-04 -2.5472E-04 6.6936E-03
41 3.4925E-04 -3.2612E-04 8.1596E-03
42 2.8678E-04 -2.8720E-04 6.4855E-03
43 1.4163E-04 -1.4926E-04 3.3112E-03
44 2.1400E-04 -2.3936E-04 4.3949E-03
45 1.2424E-04 -1.5558E-04 3.0408E-03
46 2.7340E-05 -3.5790E-05 5.9417E-04
47 7.2215E-05 -9.6257E-05 1.5233E-03
48 2.2974E-05 -3.0302E-05 4.7674E-04
49 1.5246E-04 -2.2541E-04 2.9677E-03
50 1.1097E-04 -1.8360E-04 1.7649E-03
51 2.8048E-05 -5.3701E-05 5.0807E-04
52 7.6335E-05 -1.5167E-04 1.3984E-03
53 2.9500E-04 -6.2994E-04 5.4989E-03
54 1.3718E-04 -3.1099E-04 1.9844E-03
55 3.0486E-04 -7.2088E-04 5.0718E-03
56 2.2166E-04 -5.3923E-04 2.8143E-03
57 2.8217E-04 -6.7714E-04 3.5159E-03
58 1.6164E-04 -3.3247E-04 2.1026E-03
59 3.0000E-04 -6.5640E-04 3.1621E-03
60 5.0678E-04 -1.0792E-03 4.8527E-03
61 5.7681E-05 -8.7342E-05 4.7504E-04
62 3.0594E-04 -6.2686E-04 2.0945E-03
63 2.2535E-04 -4.0760E-04 1.7729E-03
64 1.9245E-04 -3.5872E-04 1.4543E-03
65 6.6674E-05 -1.3797E-04 4.3374E-04
66 3.3064E-04 -5.1874E-04 2.2962E-03
67 2.9095E-04 -4.5419E-04 1.8807E-03
68 5.1987E-05 -7.5088E-05 3.1526E-04
69 5.8854E-04 -8.5478E-04 3.0208E-03
70 4.1099E-04 -7.1338E-04 1.7490E-03
71 8.5819E-04 -1.3405E-03 4.3646E-03
72 9.7106E-05 -2.5341E-05 2.1843E-04
73 6.3698E-04 -7.7299E-04 2.5855E-03
74 2.1654E-03 -2.0489E-03 6.4043E-03
75 2.4547E-04 -3.7235E-04 6.9292E-04
76 1.0286E-03 -5.5024E-04 1.9443E-03
77 7.9496E-04 -5.2036E-04 2.0518E-03
78 1.1633E-05 -4.1199E-04 4.3688E-04
79 4.1893E-04 -1.6219E-04 8.4874E-04
80 1.0671E-04 -6.8021E-04 7.8182E-04
81 2.3203E-03 -9.9401E-04 4.6543E-03
82 1.5328E-04 -3.1823E-05 3.2999E-04
83 2.7539E-04 -8.2324E-04 1.0520E-03
84 1.6432E-04 -7.7786E-05 6.4772E-04
85 3.3128E-04 -1.4793E-03 1.9299E-03
86 5.8071E-04 -1.2966E-04 9.6470E-04
87 7.2897E-04 -1.8707E-04 1.0648E-03
88 1.0165E-04 -6.0134E-04 9.5099E-04
89 1.9705E-04 -2.7280E-05 5.4711E-04
90 5.1470E-04 -2.8447E-05 4.2588E-04
91 4.7360E-04 -1.2936E-04 8.4947E-04
92 2.6906E-05 -1.3329E-03 1.8984E-03
93 2.5613E-04 -2.6531E-04 6.5075E-04
94 2.3538E-04 -2.1992E-05 2.0144E-04
95 1.5775E-03 -3.7759E-05 5.4441E-04
96 3.3280E-04 -7.9568E-06 1.7036E-04
97 5.9346E-04 -8.3159E-06 6.5965E-05
98 2.2511E-04 -1.5893E-05 3.1849E-04
99 2.0225E-04 -7.0445E-06 1.9922E-04
100 2.7342E-04 -1.1609E-05 3.2313E-04
101 2.6957E-04 -3.0399E-05 4.6923E-04
102 3.3244E-04 -2.7060E-05 3.2659E-04
103 1.9461E-04 -4.2923E-05 5.9301E-04
104 3.2556E-04 -5.2598E-05 5.4021E-04
105 2.3730E-04 -1.1291E-04 5.1509E-04
106 3.4607E-04 -5.8981E-05 2.2766E-04
107 1.1425E-04 -2.9850E-04 7.9441E-04
108 4.1191E-05 -6.7330E-04 1.4840E-03
109 1.9773E-04 -1.6031E-03 2.7789E-03
110 1.1533E-03 -3.4513E-04 2.8717E-04
111 4.0019E-04 -1.5092E-04 4.0941E-04
112 2.7434E-04 -5.7168E-05 2.7772E-04
113 4.5490E-04 -1.2471E-04 4.5389E-04
114 2.0751E-05 -3.1118E-05 1.8411E-04
115 1.7034E-04 -5.9649E-05 5.3270E-04
116 4.5462E-04 -9.8437E-05 8.1464E-04
117 1.0112E-03 -1.3308E-04 9.1834E-04
118 1.2441E-03 -3.4617E-04 1.8797E-03
119 1.4613E-04 -1.7858E-03 3.0512E-03
120 1.1927E-04 -1.4793E-03 3.4795E-03
121 1.6146E-03 -6.3939E-04 8.1885E-04
122 2.0288E-04 -1.4691E-04 2.7061E-04
123 4.9382E-04 -2.3581E-04 9.1055E-04
124 5.0753E-04 -9.0473E-05 4.6140E-04
125 3.2433E-04 -7.0258E-05 5.3142E-04
126 2.1587E-04 -4.8428E-05 4.5908E-04
127 7.7506E-04 -4.2735E-05 1.8533E-04
128 4.4479E-04 -5.6181E-05 4.7548E-04
129 1.3698E-03 -5.9501E-05 1.0153E-03
130 2.7637E-04 -2.5432E-05 7.1524E-04
131 5.7207E-04 -3.5591E-04 1.2909E-03
132 1.1350E-03 -1.4174E-04 7.8511E-04
133 3.6109E-03 -3.1864E-04 1.2579E-03
134 1.7268E-04 -1.4722E-03 3.9073E-03
135 2.5746E-04 -6.2532E-04 1.1511E-03
136 4.3943E-05 -1.0605E-03 2.9541E-03
137 5.7508E-06 -2.4973E-04 3.3501E-03
138 3.7473E-04 -1.2848E-03 2.3995E-03
139 2.8865E-04 -9.6386E-04 1.2706E-03
140 3.6819E-04 -1.1046E-03 2.0471E-03
141 1.4780E-04 -3.9461E-04 1.3134E-03
142 1.3985E-04 -1.8380E-04 1.0559E-03
143 1.5490E-04 -5.5020E-04 1.4747E-03
144 6.4744E-05 -1.5547E-04 7.5336E-04
145 8.1785E-04 -1.1830E-04 6.1400E-04
146 6.7829E-04 -1.7972E-04 9.0948E-04
147 3.2822E-04 -5.0617E-05 2.4536E-04
148 1.0819E-04 -2.5691E-05 1.1098E-04
149 6.4077E-05 -1.7786E-05 9.2894E-05
150 1.7080E-04 -5.5107E-05 2.8871E-04
151 1.3528E-04 -5.7736E-05 3.6300E-04
152 7.9303E-05 -5.8377E-05 3.3702E-04
153 8.0053E-05 -5.7765E-05 2.9098E-04
154 9.4301E-05 -6.1205E-05 3.5845E-04
155 9.5100E-05 -5.5160E-05 3.1008E-04
156 9.2093E-05 -4.9272E-05 3.3308E-04
157 1.1417E-04 -6.0554E-05 3.5854E-04
158 1.2913E-04 -6.2998E-05 3.6015E-04
159 3.0061E-04 -8.2329E-05 7.1490E-04
160 1.2367E-04 -4.5006E-05 2.9688E-04
161 1.4840E-04 -6.7230E-05 3.8590E-04
162 1.7126E-04 -7.3391E-05 4.2556E-04
163 1.8465E-04 -7.6909E-05 4.4451E-04
164 2.0831E-04 -8.3782E-05 4.5114E-04
165 2.2843E-04 -8.8648E-05 4.6432E-04
166 1.4190E-04 -5.2854E-05 2.4605E-04
167 1.5012E-04 -5.2102E-05 3.1876E-04
168 1.8061E-04 -5.6757E-05 3.1259E-04
169 2.1245E-04 -6.0593E-05 3.0542E-04
170 2.6343E-04 -6.3934E-05 3.4215E-04
171 1.9448E-04 -3.1752E-05 1.3489E-04
172 2.6013E-04 -3.0902E-05 1.1947E-04
173 3.7054E-04 -3.2340E-05 1.2321E-04
174 4.7482E-04 -3.2632E-05 1.1158E-04
175 2.1975E-04 -1.3672E-05 3.2459E-05
176 2.1632E-04 -1.3213E-05 4.0032E-05
177 2.2152E-04 -1.3437E-05 4.5984E-05
178 2.3849E-04 -1.4859E-05 1.8672E-05
179 2.1745E-04 -1.4007E-05 2.7954E-05
180 2.2646E-04 -1.4890E-05 3.7056E-05
181 2.1143E-04 -1.4534E-05 3.4536E-05
182 2.0606E-04 -1.4947E-05 6.3531E-05
183 1.9077E-04 -1.4574E-05 5.0864E-05
184 1.9528E-04 -1.5566E-05 4.4139E-05
185 1.8839E-04 -1.5884E-05 4.6885E-05
186 1.8455E-04 -1.6210E-05 4.8693E-05
187 1.8045E-04 -1.6486E-05 1.6260E-05
188 1.7154E-04 -1.6166E-05 4.8868E-05
189 1.7634E-04 -1.7504E-05 2.2074E-05
190 4.1297E-04 -4.3034E-05 1.3305E-04
191 4.0310E-04 -4.5888E-05 1.4039E-04
192 3.9172E-04 -4.5833E-05 1.5921E-04
193 3.8144E-04 -4.6490E-05 1.0588E-04
194 8.3313E-04 -1.0632E-04 3.3357E-04
195 8.7545E-04 -1.1477E-04 2.7002E-04
196 9.0294E-04 -1.1976E-04 3.7939E-04
197 1.0334E-03 -1.3672E-04 3.6811E-04
198 1.1556E-03 -1.4920E-04 4.2186E-04
199 6.6467E-04 -8.4875E-05 1.4783E-04
200 7.0922E-04 -8.7084E-05 2.2883E-04
201 1.6056E-03 -1.9185E-04 4.5221E-04
202 1.9989E-03 -2.2347E-04 4.2581E-04
203 2.4690E-03 -2.5230E-04 4.1739E-04
204 3.3101E-03 -3.0014E-04 0.0000E+00
205 2.0584E-03 -1.6521E-04 1.0675E-04
206 2.5065E-03 -1.8312E-04 0.0000E+00
207 2.9577E-03 -1.9366E-04 0.0000E+00
208 3.8129E-03 -2.3258E-04 -1.4373E-04
209 4.2586E-03 -2.5331E-04 9.6694E-06
210 4.8357E-03 -3.0345E-04 -1.5320E-05
211 5.4913E-03 -3.8129E-04 -2.2865E-04
212 6.4549E-03 -4.8716E-04 -3.0036E-04
213 8.8549E-03 -7.0184E-04 0.0000E+00
214 1.3538E-02 -1.0984E-03 -2.0716E-03
215 2.2933E-02 -1.8610E-03 -4.7201E-03
216 4.2566E-02 -3.3462E-03 -6.9812E-03
217 2.5852E-02 -1.9896E-03 -4.0796E-03
218 3.4808E-02 -2.6485E-03 -8.1183E-03
219 4.5763E-02 -3.4270E-03 -6.2337E-03
220 5.8912E-02 -4.3180E-03 -1.0982E-02
221 7.5765E-02 -5.4456E-03 -1.7604E-02
222 9.4178E-02 -6.7228E-03 -1.9553E-02
223 1.1300E-01 -8.0852E-03 -2.5022E-02
224 5.6986E-02 -4.0496E-03 -1.1570E-02
225 1.9220E-01 -1.3369E-02 -3.8871E-02
226 2.5046E-02 -1.7355E-03 -5.2696E-03
227 2.0587E-02 -1.4265E-03 -3.8972E-03
228 6.7126E-03 -4.6879E-04 -1.5915E-03
229 5.4068E-03 -3.7844E-04 -8.8839E-04
230 2.3059E-03 -1.6176E-04 -4.7061E-04
231 1.8324E-03 -1.2931E-04 -3.5961E-04
232 1.5083E-03 -1.0623E-04 -3.3234E-04
233 7.3805E-04 -5.2455E-05 -8.8918E-05
234 4.2877E-04 -3.0590E-05 -8.5943E-05
235 3.7055E-04 -2.6389E-05 -6.6216E-05
236 2.8675E-04 -2.0539E-05 -4.1094E-05
237 2.4347E-04 -1.7796E-05 -4.2897E-05
238 2.3790E-05 -1.7598E-06 0.0000E+00
Producing composite datafile:
6 profiles
-----------------------------------------------------------------------------------------
TSUNAMI-IP Execution Complete
----------------------------------------------------------------------------------------
6.3.1.7.3. HTML output description
The HTML formatted output from the TSUNAMI-IP sample problem is described in this section. The HTML output is generated when the html keyword is included in the PARAMETER block keyword The input for this sample problem is named tsunami-ip.input. In this case, the HTML formatted output is stored in a file called tsunami-ip.html and additional resources are stored in directories called tsunami-ip.htmd and applet_resources. These sections contain example TSUNAMI-IP HTML formatted output only for demonstration of the interface. When tsunami-ip.html is opened in a web browser, the information shown in Fig. 6.3.4 is displayed. The title of the input file is displayed between the two SCALE logos. Because this SCALE input file only executed tsunami-ip, only a single output listing is available. The text “1. TSUNAMI-IP” is a hyperlink to view the output from TSUNAMI-IP. Clicking on the “1. TSUNAMI-IP” hyperlink will present the information shown in Fig. 6.3.5
Fig. 6.3.4 Initial screen from TSUNAMI-IP HTML output.
The initial page of output from TSUNAMI-IP is shown in Fig. 6.3.5. Program verification information is shown in the table under the TSUNAMI logo. This table includes information about the code that was executed and the date and time it was run. The menu on the left side of the screen contains hyperlinks to specific portions of the code output. Echoes of the input data are available in the Input Data section. Any errors or warning messages are available in the Messages sections. Results from the code execution are shown in the results section.

Fig. 6.3.5 Program verification screen from TSUNAMI-IP HTML output.
Selecting Input Parameters will reveal the menu of available input data. Selecting Input Parameters causes the table shown in Fig. 6.3.6 to be displayed. Other input data can also be displayed by selecting the desired data from the menu.
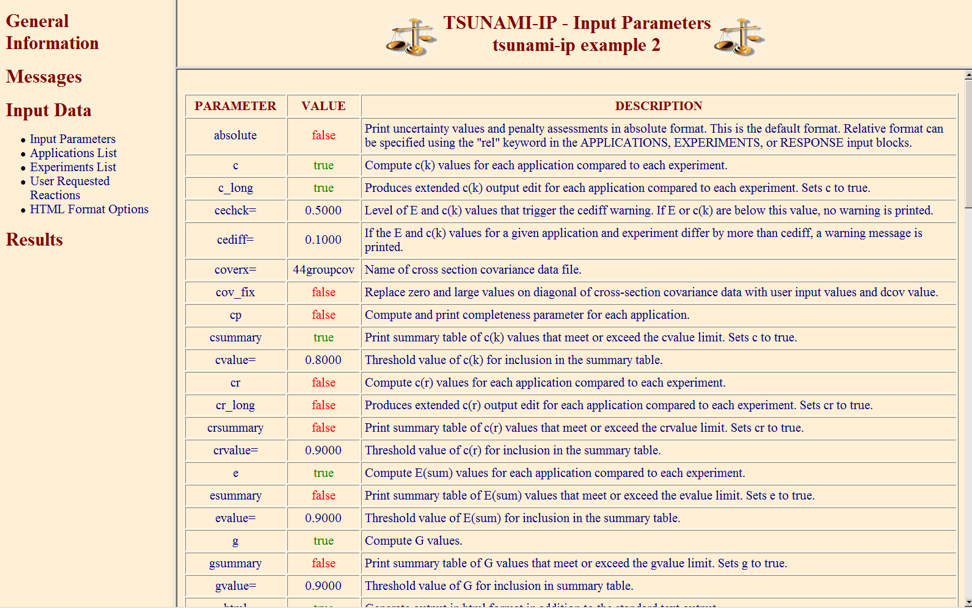
Fig. 6.3.6 Input parameters from TSUNAMI-IP HTML output.
Selecting Messages will reveal a menu of available messages. Selecting Warning Messages from the Messages section of the menu causes the information shown in Fig. 6.3.7 to appear. The Warning Messages edit contains all warning messages that were generated during the execution of the code. If errors were encountered in the code execution, an Error Messages item would have also been available in the menu under Messages.
Fig. 6.3.7 Warning messages from TSUNAMI-IP HTML output.
Selecting Results causes a menu of available results to be revealed. From this menu, selecting Global Integral Indices causes a submenu of available global integral indices to be revealed. Selecting Integral Index Values from this submenu causes the information shown in Fig. 6.3.8 to appear. In certain results edits, a second menu appears on the right such that the information for a particular application can be quickly reached. The table shown in Fig. 6.3.8 corresponds to the values table from the standard output file. One advantage of the HTML output is the use of color coding. Values exceeding the cutoff value for a particular integral index are printed in the color that is set by the HMTL data block input cut_clr=. The maximum value for each index for each application is printed in the color that is set by the HMTL data block input max_clr=. In this case, the values that exceed the cutoff values are also the maximum values and are colored as such. Edits for each type of data requested in the PARAMETER data block are available in the results section by selecting the appropriate submenu.
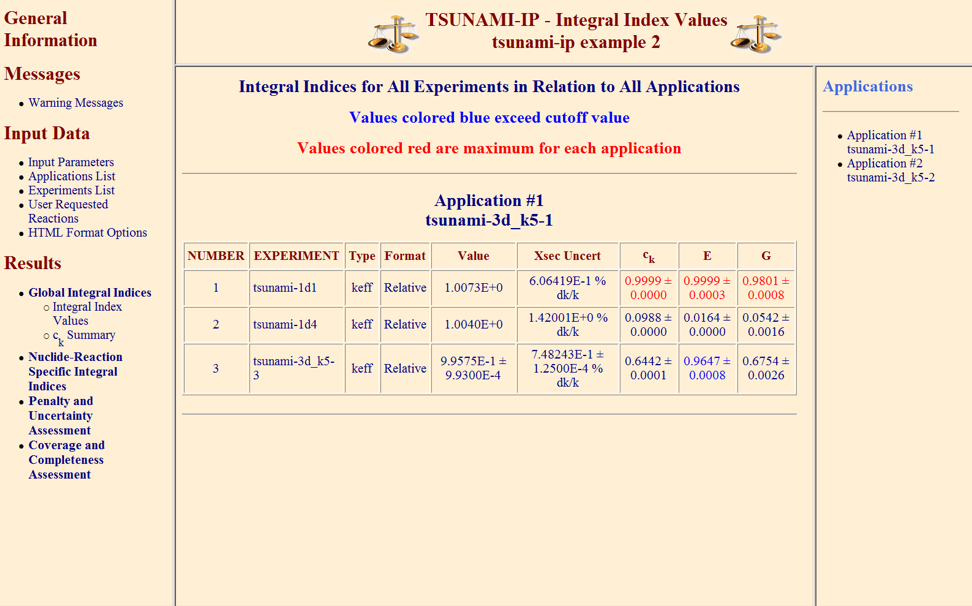
Fig. 6.3.8 Global integral indices from TSUNAMI-IP HTML output.
Data plots can also be directly viewed in the HTML output. The composite data can be viewed by selecting Coverage and Completeness Assessment then Composite Sensitivity Data Plot. A Java applet version of Javapeño will appear in the browser window with the appropriate datafile preloaded. Data can be added to the plot by double-clicking on the list of available data on the right side of Javapeño. The plot shown in Fig. 6.3.9 was produced with this procedure.
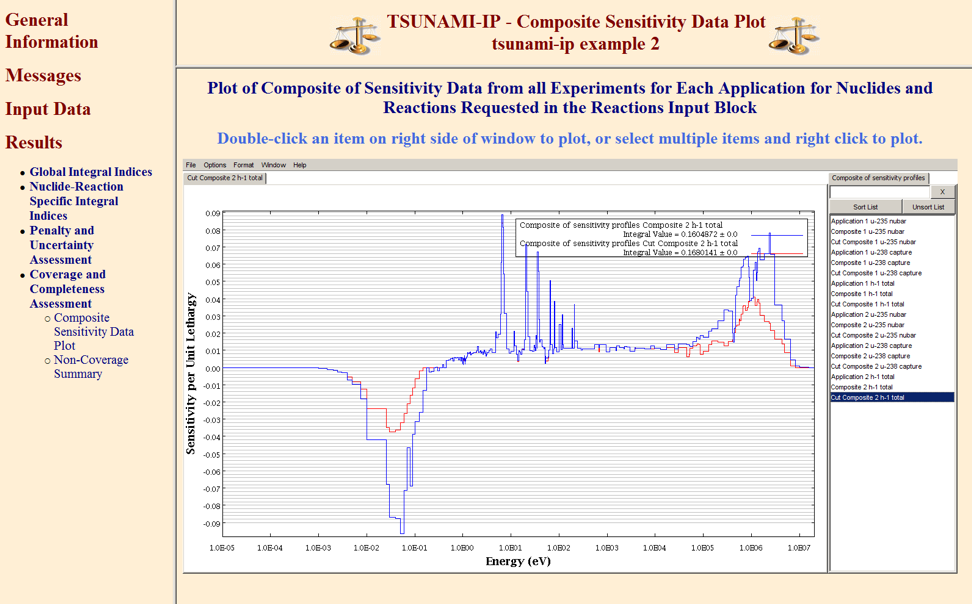
Fig. 6.3.9 Composite sensitivity data plot from TSUNAMI-IP HTML output.
6.3.2. BONAMIST
BONAMIST is a sensitivity version of the SCALE 4.4a BONAMI module. BONAMI performs Bondarenko calculations for resonance self-shielding. The entire FORTRAN 77 source for BONAMI was processed with the GRESS pre-compiler so that the sensitivities of the self-shielded cross sections to the data input to the code are computed. BONAMIST utilizes many of the routines from SCALELIB. BONAMIST writes the resonance self-shielded cross sections to an AMPX formatted data file, just as BONAMI does. However, an additional data file, bonamist.sen, is also written. The bonamist.sen file contains the sensitivities of resonance self-shielded cross sections generated by BONAMIST to the quantities input to BONAMIST.
The methodologies for computing the resonance self-shielded cross sections in BONAMIST are identical to those used in BONAMI, and equivalent cross section results are obtained. The bonamist.sen data file contains the sensitivity of the self-shielded cross sections to the number density of each nuclide and the extra cross section input to BONAMIST. The extra cross section is used by TSUNAMI-1D, TSUNAMI-3D, CSAS and other SCALE sequences to pass the Dancoff factor computed in MIPLIB or SENLIB to BONAMI. The format of the bonamist.sen data file is given in Format of BONAMIST.SEN data file.
The input, output, methodologies and program flow of BONAMIST are not described here. The user is referred to the BONAMI chapter. The input of BONAMIST is identical to that required for BONAMI. The sensitivities are automatically computed and the bonamist.sen data file is automatically generated without user intervention.
6.3.3. References
- refTsip1(1,2)
B. L. Broadhead, B. T. Rearden, C. M. Hopper, J. J. Wagschal, and C. V. Parks, “Sensitivity- and Uncertainty-Based Criticality Safety Validation Techniques,” Nucl. Sci. Eng., 146, 340–366 (2004).
- refTsip2(1,2)
S. Golouglu, C. M. Hopper, and B. T. Rearden, “Extended Interpretation of Sensitivity Data for Benchmark Areas of Applicability,” Trans. Am. Nuc. Soc., 88, 77–79 (2003).
- refTsip3(1,2)
S. Goluoglu, K. R. Elam, B. T. Rearden, B. L. Broadhead, and C. M. Hopper, Sensitivity Analysis Applied to the Validation of the 10B Capture Reaction in Nuclear Fuel Casks, NUREG/CR-6845 (ORNL/TM-2004/48) U.S. Nuclear Regulatory Commission, Oak Ridge National Laboratory, August 2004.
- refTsip4
B. L. Broadhead, C. M. Hopper, K. R. Elam, B. T. Rearden, and R. L. Childs, “Criticality Safety Applications of S/U Validation Methods,” Trans. Am. Nucl. Soc. 83, 107–113 (2000).
- refTsip5
M. E. Dunn, PUFF-III: A Code for Processing ENDF Uncertainty Data Into Multigroup Covariance Matrices, ORNL/TM-1999/235 (NUREG/CR-6650), U.S. Nuclear Regulatory Commission, Oak Ridge National Laboratory, June 2000.
- refTsip6
J. J. Lichtenwalter, S. M. Bowman, M. D. DeHart, and C. M. Hopper, Criticality Benchmark Guide for Light-Water-Reactor Fuel in Transportation and Storage Packages, NUREG/CR-6361 (ORNL/TM-13211), U.S. Nuclear Regulatory Commission, Oak Ridge National Laboratory, March 1997.